The Roles of MicroRNAs in the Regulation of Rice–Pathogen Interactions
Abstract
:1. Introduction
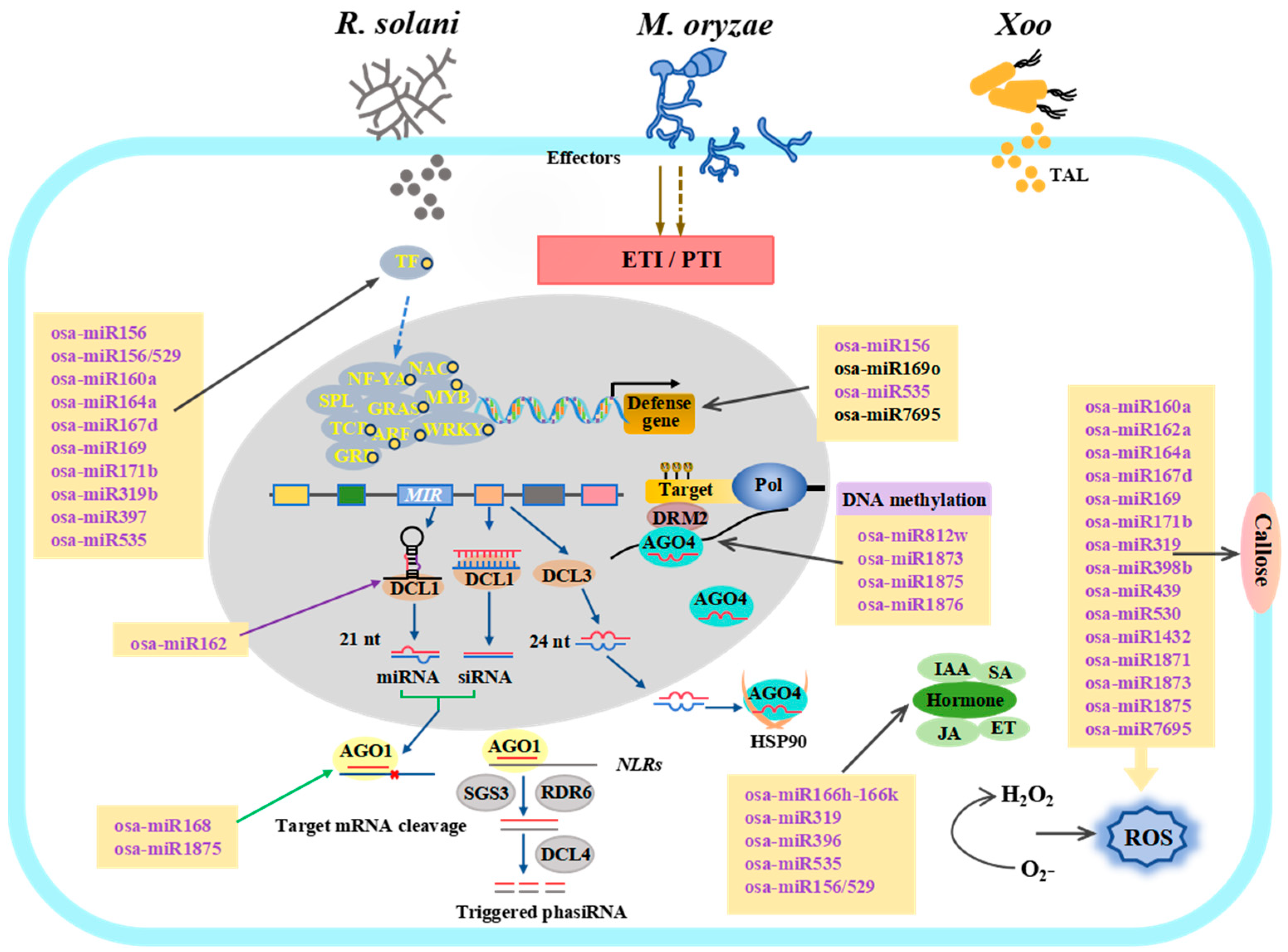
2. Functionally Characterized Osa-miRNAs Associated with the Immune Response Against M. oryzae, Xoo, and R. solani
3. Identification of Osa-miRNAs Involved in Response to M. oryzae in Rice
3.1. The Osa-miRNAs That Play Positive Roles in Rice Immunity Against M. oryzae
3.2. The Osa-miRNAs That Facilitate M. oryzae Attack in Rice
4. Identification of RiceOsa-miRNAs Responsive to Xoo
4.1. The Osa-miRNAs That Play Positive Roles in Rice Immunity Against Xoo
4.2. The Osa-miRNAs That Facilitate Xoo Attack in Rice
5. Identification of Rice Osa-miRNAs Responsive to R. solani
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Mencia, R.; Gonzalo, L.; Tossolini, I.; Manavella, P.A. Keeping up with the miRNAs: Current paradigms of the biogenesis pathway. J. Exp. Bot. 2023, 74, 2213–2227. [Google Scholar] [CrossRef]
- Shang, R.F.; Lee, S.; Senavirathne, G.; Lai, E.C. MicroRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Chang, L.; Qi, Y. DNA methylation mediated by a MicroRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef]
- Huang, T.X.; Li, Y.; Wang, W.; Xu, L.; Li, J.R.; Yijun Qi, Y.J. Evolution of lmiRNAs and their targets from MITEs for rice adaptation. J. Integr. Plant Biol. 2022, 64, 2411–2424. [Google Scholar] [CrossRef]
- Tang, J.Y.; Chu, C.C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Dong, Q.K.; Hu, B.B.; Zhang, C. MicroRNAs and their roles in plant development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.; Jaiswal, S.; Iquebal, M.A.; Singh, R.; Gautam, R.K.; Rai, A.; Kumar, D. Revitalizing miRNAs mediated agronomical advantageous traits improvement in rice. Plant Physiol. Biochem. 2023, 202, 107933. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Charagh, S.; Karikari, B.; Sharif, R.; Yadav, V.; Mubarik, M.S.; Habib, M.; Zhuang, Y.H.; Zhang, C.; Chen, H.; et al. MiRNAs for crop improvement. Plant Physiol. Biochem. 2023, 201, 107857. [Google Scholar] [CrossRef] [PubMed]
- Asadi, M.; Millar, A.A. Review: Plant microRNAs in pathogen defense: A panacea or a piece of the puzzle? Plant Sci. 2024, 341, 111993. [Google Scholar] [CrossRef]
- Shen, E.H.; Zhao, T.L.; Zhu, Q.H. Are miRNAs applicable for balancing crop growth and defense trade-off? New Phytol. 2024, 243, 1670–1680. [Google Scholar] [CrossRef]
- Zhan, J.P.; Meyers, B.C. Plant small RNAs: Their biogenesis, regulatory roles, and functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, L.; Tossolini, I.; Gulanicz, T.; Cambiagno, D.A.; Kasprowicz-Maluski, A.; Smolinski, D.J.; Mammarella, M.F.; Ariel, F.D.; Marquardt, S.; Szweykowska-Kulinska, Z.; et al. R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants. Nat. Plants 2022, 8, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jia, T.R.; Chen, X.M. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef]
- Li, S.B.; Le, B.; Ma, X.; Li, S.F.; You, C.J.; Yu, Y.; Zhang, B.L.; Liu, L.; Gao, L.; Shi, T.; et al. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 2016, 5, e22750. [Google Scholar] [CrossRef]
- Xu, Y.; Xuemei Chen, X.M. MicroRNA biogenesis and stabilization in plants. Fundam. Res. 2023, 3, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, Q.; Zhou, H.; Ni, F.; Wu, X.; Qi, Y. Rice microRNA effector complexes and targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.N.; Sun, Z.H.; Li, H.Y.; Liu, H. The biosynthesis process of small RNA and its pivotal roles in plant development. Int. J. Mol. Sci. 2024, 25, 7680. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xue, Y.; Zhang, L.X.; Zhong, Z.H.; Feng, S.H.; Wang, C.S.; Xiao, L.F.; Yang, Z.L.; Harris, C.J.; Wu, Z.; et al. Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation. Science 2021, 374, 1152–1157. [Google Scholar] [CrossRef]
- Yuan, S.; Linquist, B.A.; Wilson, L.T.; Cassman, K.G.; Stuart, A.M.; Pede, V.; Miro, B.; Saito, K.; Agustiani, N.; Aristya, V.E.; et al. Sustainable intensification for a larger global rice bowl. Nat. Commun. 2021, 12, 7163. [Google Scholar] [CrossRef]
- Dai, L.Y.; Liu, X.L.; Xiao, Y.H.; Wang, G.L. Recent advances in cloning and characterization of disease resistance genes in rice. J. Integr. Plant Biol. 2010, 49, 112–119. [Google Scholar] [CrossRef]
- Lee, F.N.; Rush, M.C. Rice sheath blight: A major rice disease. Plant Dis. 1983, 67, 829–832. [Google Scholar] [CrossRef]
- Mizukami, T.; Wakimoto, S. Epidemiology and control of bacterial leaf blight of rice. Annu. Rev. Phytopathol. 1969, 7, 51–72. [Google Scholar] [CrossRef]
- Ou, S.H. Pathogen variability and host resistance in rice blast disease. Annu. Rev. Phytopathol. 1980, 18, 167–187. [Google Scholar] [CrossRef]
- Esse, H.P.; Reuber, T.L.; Does, D.V. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhu, L.; He, G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant 2013, 6, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Ning, Y.; Yang, D.L.; Zhai, K.; Wang, G.L.; He, Z. Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol. Plant 2020, 13, 1402–1419. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef]
- Kourelis, J.; Hoorn, R.A.L. Defended to the Nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.L. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.Z.; Yang, Z.R.; Yang, R.X.; Huang, Y.; Guo, G.; Kong, X.Y.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.M.; et al. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant. 2019, 12, 1114–1122. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, S.; Yu, B. MicroRNA biogenesis, degradation and activity in plants. Cell Mol. Life Sci. 2015, 72, 87–99. [Google Scholar] [CrossRef]
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.C.; Segundo, B.S. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jeyakumar, J.M.J.; Feng, Q.; Zhao, Z.; Fan, J.; Khaskheli, M.I.; Wang, W. The roles of rice microRNAs in rice Magnaporthe oryzae interaction. Phytopathol. Res. 2019, 1, 33. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, S.; Li, J.; Hu, X.; Wang, H.; Cao, X.; Xu, Y.; Zhao, Z.; Xiao, Z.; Yang, N.; et al. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Feng, Q.; Cao, X.; Zhu, Y.; Wang, H.; Chandran, V.; Fan, J.; Zhao, J.; Pu, M.; Li, Y.; et al. Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol. 2020, 62, 702–715. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; He, X.; Zhu, Y.; Li, T.; Lin, X.; Mao, W.; Gishkori, Z.G.N.; Zhao, Z.; Zhang, J.; et al. The rice miR171b–SCL6-IIs module controls blast resistance, grain yield, and flowering. Crop J. 2022, 10, 117–127. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.; Zheng, Y.; Liu, X.; Zhou, S.; Yang, X.; Liu, S.; Li, Y.; Li, J.; Zhao, S.; et al. Osa-miR535 targets SQUAMOSA promoter binding protein-like 4 to regulate blast disease resistance in rice. Plant J. 2022, 110, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Wang, H.; Yang, X.; Hu, Z.W.; Zhou, X.H.; Xiang, L.; Xiong, X.Y.; He, X.R.; Zhu, Y.; Li, G.B.; et al. Osa-miR160a confers broad-spectrum resistance to fungal and bacterial pathogens in rice. New Phytol. 2022, 236, 2216–2232. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, Y.; Shi, Y.; Wu, L.; Xu, Y.; Huang, F.; Guo, X.; Zhang, Y.; Fan, J.; Zhao, J.; et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.Y.; Feng, R.Y.; Chen, S.L. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Zheng, Y.; Wang, H.; Yang, X.; Chen, J.; Zhou, S.; Wang, L.; Li, X.; Ma, X.; et al. Expressing a target mimic of miR156fhl-3p enhances rice blast disease resistance without yield penalty by improving SPL14 expression. Front. Genet. 2020, 11, 327. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.; Zhu, Y.; Yang, X.; Zhang, K.; Xiao, Z.; Wang, H.; Zhao, J.; Zhang, L.; Li, G.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Sanuy, F.; Peris-Peris, C.; Tomiyama, S.; Okada, K.; Hsing, Y.I.; Segundo, B.S.; Campo, S. Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol. 2019, 19, 563. [Google Scholar] [CrossRef]
- Campo, S.; Peris-Peris, C.; Sire, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; Segundo, B.S. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Quoc, N.B.; Phuong, N.D.N.; Trang, H.T.T.; Phi, N.B.; Chau, N.N.B. Expression of osa-miR7695 against the blast fungus Magnaporthe oryzae in vietnamese rice cultivars. Eur. J. Plant Pathol. 2019, 155, 307–317. [Google Scholar] [CrossRef]
- Salvador-Guirao, R.; Hsing, Y.I.; Segundo, B.S. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front. Plant Sci. 2018, 9, 337. [Google Scholar] [CrossRef]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.; Chen, Y.; Li, G.; Zhu, Y.; Yang, X.M.; Zhang, L.L.; Zhao, Z.X.; et al. miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 2018, 9, 1999. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, X.; Wang, H.; Zhu, Y.; Liu, X.; Li, T.; Zheng, Y.; Zhao, J.; Zhang, J.; Huang, Y.; et al. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and yield. Rice 2020, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.; Lu, J.; Li, X.; Dang, W.; Ma, X.; Yang, Z.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Li, X.; Xia, S.; Zhang, Y.; Yu, Z.; Tang, C.; Xu, L.; Wang, Z.; Zhang, X.; Zhou, T.; et al. An OsPRMT5-OsAGO2/miR1875-OsHXK1 module regulates rice immunity to blast disease. J. Integr. Plant Biol. 2023, 65, 1077–1095. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Sanchez-Sanuy, F.; Camargo-Ramırez, R.; Gomez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suarez, M.; Segundo, B.S. A novel transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, Y.; Wang, L.; Zheng, Y.; Chen, J.; Li, T.; Yang, X.; Wang, H.; Li, X.; Ma, X.; et al. Osa-miR1873 fine-tunes rice immunity against Magnaporthe oryzae and yield traits. J. Integr. Plant Biol. 2020, 62, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, Z.; Li, Y.; Li, T.; Zhu, Y.; Yang, X.; Zhou, S.; Wang, H.; Zhao, J.; Pu, M.; et al. Fine-tuning roles of Osa-miR159a in rice immunity against Magnaporthe oryzae and development. Rice 2021, 14, 26. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.; He, X.; Zhu, Y.; Feng, Q.; Yang, X.M.; Zhou, X.H.; Li, G.; Ji, Y.; Zhao, J.; et al. Blocking Osa-miR1871 enhances rice resistance against Magnaporthe oryzae and yield. Plant Biotechnol. J. 2022, 20, 646–659. [Google Scholar] [CrossRef]
- Lu, J.; Yang, X.; Chen, J.; Li, T.; Hu, Z.; Xie, Y.; Li, J.; Zhao, J.; Pu, M.; Hui, F.; et al. Osa-miR439 negatively regulates rice immunity against Magnaporthe oryzae. Rice Sci. 2021, 28, 156–165. [Google Scholar]
- Li, Y.; Wang, L.; Bhutto, S.; He, X.; Yang, X.; Zhou, X.; Lin, X.; Rajput, A.; Li, G.; Zhao, J.; et al. Blocking miR530 improves rice resistance, yield, and maturity. Front. Plant Sci. 2021, 12, 729560. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Zhou, X.; Yang, X.; He, X.; Feng, Q.; Zhu, Y.; Li, G.B.; Wang, H.; Zhao, J.; et al. Rice miR1432 fine-tunes the balance of yield and blast disease resistance via different modules. Rice 2021, 14, 87. [Google Scholar] [CrossRef]
- Liu, M.; Shi, Z.; Zhang, X.; Wang, M.; Zhang, L.; Zheng, K.; Liu, J.; Hu, X.; Di, C.; Qian, Q.; et al. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 2019, 5, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Ke, Y.; Chen, D.; Wang, L.; Li, Q.; Yuan, M. Rice microRNA156/529-SQUAMOSA PROMOTER BINDING modules regulate defenses against bacteria. Plant Physiol. 2023, 192, 2537–2553. [Google Scholar] [CrossRef]
- Yu, C.; Chen, Y.; Cao, Y.; Chen, H.; Wang, J.; Bi, Y.; Tian, F.; Yang, F.; Rothstein, S.J.; Zhou, X.; et al. Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol. 2018, 59, 1234–1247. [Google Scholar] [CrossRef]
- Jiang, G.; Liu, D.; Yin, D.; Zhou, Z.; Shi, Y.; Li, C.; Zhu, L.; Zhai, W. A rice NBS-ARC gene conferring quantitative resistance to bacterial blight is regulated by a pathogen effector-inducible miRNA. Mol. Plant 2020, 13, 1752–1767. [Google Scholar] [CrossRef]
- Jia, Y.; Li, C.; Li, Q.; Liu, P.; Liu, D.; Liu, Z.; Wang, Y.; Jiang, G.; Zhai, W. Characteristic dissection of Xanthomonas oryzae pv. oryzae responsive microRNAs in rice. Int. J. Mol. Sci. 2020, 21, 785. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Li, Q.; Li, Y.; Zhai, W.; Jiang, G.; Li, C. Inducible enrichment of osa-miR1432 confers rice bacterial blight resistance through suppressing OsCaML2. Int. J. Mol. Sci. 2021, 22, 11367. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hui, S.; Lv, Y.; Zhang, M.; Chen, D.; Tian, J.; Zhang, H.; Liu, H.; Cao, J.; Xie, W.; et al. miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol. Plant 2022, 15, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kuang, Y.; Chen, Y.; Shi, J.; Cao, Y.; Hu, J.; Yu, C.; Yang, F.; Tian, F.; Chen, H. miR2118 negatively regulates bacterial blight resistance through targeting several disease resistance genes in rice. Plants 2023, 12, 3815. [Google Scholar] [CrossRef] [PubMed]
- Srikakulam, N.; Guria, A.; Karanthamalai, J.; Murugesan, V.; Krishnan, V.; Sundaramoorthy, K.; Saha, S.; Singh, R.; Victorathisayam, T.; Rajapriya, V.; et al. An insight into pentatricopeptide-mediated chloroplast necrosis via microRNA395a during Rhizoctonia solani infection. Front. Genet. 2022, 3, 869465. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Zhang, Z.; Gao, P.; Feng, Z.; Zuo, S.; Ouyang, S. Suppression of rice osa-miR444.2 improves the resistance to sheath blight in rice mediating through the phytohormone pathway. Int. J. Mol. Sci. 2023, 24, 3653. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Wu, H.; Ma, Y.; Chen, T.; Wang, M.; Wang, X. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.J.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y.; et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017, 8, 1598. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhong, Z.; Chen, H.; Li, Q.; Zheng, X.; Qi, Y.; Zhang, Y. Knocking out microRNA genes in rice with CRISPR-Cas9. Methods Mol. Biol. 2019, 1917, 109–119. [Google Scholar]
- Bi, H.; Fei, Q.; Li, R.; Liu, B.; Xia, R.; Char, S.N.; Meyers, B.C.; Yang, B. Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol. J. 2020, 18, 1526–1536. [Google Scholar] [CrossRef]
- Kumar, K.; Mandal, S.N.; Neelam, K.; Reyes, B.G. MicroRNA-mediated host defense mechanisms against pathogens and herbivores in rice: Balancing gains from genetic resistance with trade-offs to productivity potential. BMC Plant Biol. 2022, 22, 351. [Google Scholar] [CrossRef]
- Chen, X.; Ronald, P.C. Innate immunity in rice. Trends Plant Sci. 2011, 16, 451–459. [Google Scholar] [CrossRef]
- Thordal-Christensen, H. A holistic view on plant effector-triggered immunity presented as an iceberg model. Cell. Mol. Life Sci. 2020, 77, 3963–3976. [Google Scholar] [CrossRef]
- Xin, X.; Kvitko, B.; He, S. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of jasmonic acid-mediated defense by viralinducible microRNA319 facilitates virus infection in rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 21, 980–986. [Google Scholar] [CrossRef]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef]
- Wang, B.; Ebbole, D.; Wang, Z. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Xie, Z.; Yan, B.; Shou, J.; Tang, J.; Wang, X.; Zhai, K.; Liu, J.; Li, Q.; Luo, M.; Deng, Y.; et al. A nucleotide-binding siteleucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, S.; Yuan, M. An update on molecular mechanism of disease resistance genes and their application for genetic improvement of rice. Mol. Breed. 2019, 39, 154. [Google Scholar] [CrossRef]
- Dong, S.; Zhang, J.; Sun, D.; Liu, H.; Yang, Q.; Wang, H.; Chen, Z.; Wang, J. Identification of Magnaporthe oryzae-elicited rice novel miRNAs and their targets by miRNA and degradome sequencing. Eur. J. Plant Pathol. 2018, 151, 629–647. [Google Scholar] [CrossRef]
- Li, Z.; Xia, J.; Chen, Z.; Yu, Y.; Li, Q.; Zhang, Y.; Zhang, J.; Wang, C.; Zhu, X.; Zhang, W.; et al. Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci. Rep. 2016, 6, 25493. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.; Reddy, B.; Sheoran, N.; Ganesan, P.; Kumar, A. Unraveling the transcriptional network regulated by miRNAs in blast-resistant and blast-susceptible rice genotypes during Magnaporthe oryzae interaction. Gene 2023, 886, 147718. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Wang, W.; Zhang, Z.; Liang, Y.; Gao, X.; Li, P.; Liu, Y.; Zhang, L.H.; Chu, C. Identification of microRNAs in rice root in response to nitrate and ammonium. J. Genet. Genom. 2016, 43, 651–661. [Google Scholar] [CrossRef]
- Khan, A.; Goswami, K.; Sopory, S.K.; Sanan-Mishra, N. “Mirador” on the potential role of miRNAs in synergy of light and heat networks. Indian J. Plant Physiol. 2017, 22, 587–607. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Zhao, J.; Wang, S.; Jin, Y.; Chen, Z.; Fang, Y.; Hua, C.; Ding, S.; Guo, H. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Q.; Zhao, Y.; Yang, J.; He, H.; Jia, G. The lre-miR159a-LrGAMYB pathway mediates resistance to grey mould infection in Lilium regale. Mol. Plant Pathol. 2020, 21, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sahu, P.; Prasad, A.; Muthamilarasan, M.; Waseem, M.; Khan, Y.; Thakur, J.K.; Chakraborty, S.; Prasad, S. The Sw5a gene confers resistance to ToLCNDV and triggers an HR response after direct AC4 effector recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2101833118. [Google Scholar] [CrossRef]
- Yu, X.; Lin, X.; Zhou, T.; Cao, L.; Hu, K.; Li, F.; Qu, S. Host-induced gene silencing in wild apple germplasm Malus hupehensis confers resistance to the fungal pathogen Botryosphaeria dothidea. Plant J. 2024, 118, 1174–1193. [Google Scholar] [CrossRef]
- Tong, A.; Yuan, Q.; Wang, S.; Peng, J.; Lu, Y.; Zheng, H.; Lin, L.; Chen, H.; Gong, Y.; Chen, J.; et al. Altered accumulation of osa-miR171b contributes to rice stripe virus infection by regulating disease symptoms. J. Exp. Bot. 2017, 68, 4357–4367. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Zhang, Z.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.P. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, X.; Cai, W.; Huang, W.; Zhou, X.; Luo, Q.; Yang, H.; Wang, J.; Huang, J. Arabidopsis miR171-targeted scarecrow-like proteins bind to GT ciselements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet. 2014, 10, e1004519. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Hu, Y.; Liu, H.; He, M.; Yang, Z.; Kong, F.; Liu, X.; Hou, X. DELLA and EDS1 form a feedback regulatory module to fine-tune plant growth-defense tradeoff in Arabidopsis. Mol. Plant 2019, 12, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Balyan, S.; Kumar, M.; Mutum, R.D.; Raghuvanshi, U.; Agarwal, P.; Mathur, S.; Raghuvanshi, S. Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci. Rep. 2017, 7, 15446. [Google Scholar] [CrossRef]
- Ryu, M.; Mishra, R.C.; Jeon, J.; Lee, S.K.; Bae, H. Drought-induced susceptibility for Cenangium ferruginosum leads to progression of Cenangium-dieback disease in Pinus koraiensis. Sci. Rep. 2018, 8, 16368. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Shen, Y.; Li, H.; Yang, J.; Cai, X.; Zheng, G.; Zhu, Y.; Jia, B.; Sun, X. The multiple roles of OsmiR535 in modulating plant height, panicle branching and grain shape. Plant Sci. 2019, 283, 60–69. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Jin, J.; Fu, D.; Yang, X.; Weng, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 15504–15509. [Google Scholar] [CrossRef]
- Yan, Y.; Wei, M.X.; Li, Y.; Tao, H.; Wu, H.Y.; Chen, Z.F.; Li, C.; Xu, J.H. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.). Plant Sci. 2021, 302, 110728. [Google Scholar] [CrossRef]
- Wang, L.; Ming, L.; Liao, K.; Xia, C.; Sun, S.; Chang, Y.; Wang, H.; Fu, D.; Xu, C.; Wang, Z.; et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant 2021, 14, 1168–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Y.; Qin, T.; Guo, X.; Xu, K.; Xu, C.; Yuan, W. Functional conservation and divergence of miR156 and miR529 during rice development. Crop J. 2023, 11, 692–703. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 2020, 28, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, D.; Sun, F.; Guo, W.; Wang, W.; Li, X.; Lan, Y.; Du, L.; Li, S.; Fan, Y.; et al. ARGONAUTE 2 increases rice susceptibility to rice black-streaked dwarf virus infection by epigenetically regulating HEXOKINASE 1 expression. Mol. Plant Pathol. 2021, 22, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Alvarado, G.P.; Sanchez-Nieto, S. Plant hexokinases are multifaceted proteins. Plant Cell Physiol. 2017, 58, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Sun, C.; Zhang, Q.; An, J.P.; You, C.; Hao, Y. Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 2016, 12, e1006273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, S.; Wang, Z.; He, L.; Xu, K.; Wang, G. Glucose triggers stomatal closure mediated by basal signaling through HXK1 and PYR/RCAR receptors in Arabidopsis. J. Exp. Bot. 2018, 69, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Munemasa, S.; Shinya, T.; Wu, W.; Ma, T.; Lu, J.; Kinoshita, T.; Kaku, H.; Shibuya, N.; Murata, Y. Stomatal immunity against fungal invasion comprises not only chitin-induced stomatal closure but also chitosan-induced guard cell death. Proc. Natl. Acad. Sci. USA 2020, 117, 20932–20942. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Huang, J.; Yang, L.; Wang, Z.; Wu, D.; Shu, X. MOS1 negatively regulates sugar responses and anthocyanin biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7095. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Zhou, Y.; Zhao, Y.; Song, Z.; Wang, S.; Fan, J.; Hu, B.; Venturi, V.; Liu, F. Proteomic analysis reveals novel extracellular virulence-associated proteins and functions regulated by the diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzicola. J. Proteome Res. 2013, 12, 3327–3341. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Xu, R.; Liu, F.; Hu, B.; Walcott, R.R. Simultaneous detection of Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola in rice seed using a padlock probe-based assay. Phytopathology 2014, 104, 1130–1137. [Google Scholar] [CrossRef]
- Nino-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant. Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Yamanouchi, U.; Katayose, Y.; Toki, S.; Wang, Z.; Kono, I.; Kurata, N.; Yano, M.; Sasaki, T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 1998, 95, 1663–1668. [Google Scholar] [CrossRef]
- Ji, C.; Ji, Z.; Liu, B.; Cheng, H.; Liu, H.; Liu, S.; Yang, B.; Chen, G. Xa1 allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Commun. 2020, 1, 100087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, H.; Li, F.; Ouyang, Y..; Yuan, M.; Li, X.; Xiao, J.; Wang, S. Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun. 2020, 1, 100088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Coaker, G.; Zhou, J.; Dong, X. Plant immune mechanisms: From reductionistic to holistic points of view. Mol. Plant 2020, 13, 1358–1378. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cao, Y.; Yang, Z.; Xu, C.; Li, X.; Wang, S.; Zhang, Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004, 37, 517–527. [Google Scholar] [CrossRef]
- Xiang, Y.; Cao, Y.; Xu, C.; Li, X.; Wang, S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinaselike protein, is the same as Xa26. Theor. Appl. Genet. 2006, 113, 1347–1355. [Google Scholar] [CrossRef]
- Hu, K.; Cao, J.; Zhang, J.; Xia, F.; Ke, Y.; Zhang, H.; Xie, W.; Liu, H.; Cui, Y.; Cao, Y.; et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.S.; McCouch, S.R. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 2004, 17, 1348–1354. [Google Scholar] [CrossRef]
- Jiang, G.; Xia, Z.; Zhou, Y.; Wan, J.; Li, D.; Chen, R.; Zhai, W.; Zhu, L. Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAg1. Mol. Genet. Genom. 2006, 275, 354–366. [Google Scholar] [CrossRef]
- Tian, D.; Wang, J.; Zeng, X.; Gu, K.; Qiu, C.; Yang, X.; Zhou, Z.; Goh, M.; Luo, Y.; Murata-Hori, M.; et al. The rice TAL effector dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 2014, 26, 497–515. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci.USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, G.; Chen, L.; Kim, H.S.; Pi, L.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.; Zhu, L.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, X.; Fan, Y.; Gao, Y.; Zhu, Q.; Zheng, C.; Qin, T.; Li, Y.; Che, J.; Zhang, M.; et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 2015, 8, 290–302. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, M.; Zhou, Y.; Li, X.; Xiao, J.; Wang, S. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 2011, 34, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.; Yang, B.; Tian, D.; Wu, L.; Wang, D.; Sreekala, C.; Yang, F.; Chu, Z.; Wang, G.; White, F.F.; et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005, 435, 1122–1125. [Google Scholar] [CrossRef]
- Hutin, M.; Sabot, F.O.; Ghesquiere, A.; Koebnik, R.; Szurek, B. A knowledge-based molecular screen uncovers a broadspectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 2016, 84, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, P.; Mei, L.; He, X.; Chen, L.; Liu, H.; Shen, S.; Ji, Z.; Zheng, X.; Zhang, Y.; et al. Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice. Plant Commun. 2021, 2, 100143. [Google Scholar] [CrossRef]
- White, F.F.; Yang, B. Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 2009, 150, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Nie, L.X.; Khan, F.A.; Chen, Y.; Hussain, S.; Wu, C.; Xiong, D.; Jing, W.; Saud, S.; Khan, F.A.; et al. Disease resistance in rice and the role of molecular breeding in protecting rice crops against diseases. Biotechnol. Lett. 2014, 36, 1407–1420. [Google Scholar] [CrossRef] [PubMed]
- Hop, D.V.; Hoa, P.T.; Quang, N.; Ton, P.H.; Ha, T.H.; Huang, N.V.; Van, N.T.; Hai, T.V.; Quy, N.T.; Dao, N.T.; et al. Biological control of Xanthomonas oryzae pv. oryzae causing rice bacterial blight disease by Streptomyces toxytricini VN08-A-12, isolated from soil and leaf-litter samples in Vietnam. Biocontrol Sci. 2014, 19, 103–111. [Google Scholar] [CrossRef]
- Bimolata, W.; Kumar, A.; Reddy, S.K.; Sundaram, R.M.; Laha, G.S.; Qureshi, I.A.; Ghazi, I.A. Nucleotide diversity analysis of three major bacterial blight resistance genes in rice. PLoS ONE 2015, 10, e0120186. [Google Scholar] [CrossRef]
- Ji, Z.; Ji, C.; Liu, B.; Zou, L.; Chen, G.; Yang, B. Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 2016, 7, 13435. [Google Scholar] [CrossRef]
- Li, Q.; Wang, B.; Yu, J.; Dou, D. Pathogen-informed breeding for crop disease resistance. J. Integr. Plant Biol. 2021, 63, 305–311. [Google Scholar] [CrossRef]
- Dilla-Ermita, C.J.; Tandayu, E.; Juanillas, V.M.; Detras, J.; Lozada, D.N.; Dwiyanti, M.S.; Cruz, C.V.; Mbanjo, E.G.; Ardales, E.; Diaz, M.G.; et al. Genome-wide association analysis tracks bacterial leaf blight resistance loci in rice diverse germplasm. Rice 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Reinke, R.F. A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 2019, 14, e0211775. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Ji, C.H.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Li, J.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 2019, 112, 1434–1446. [Google Scholar] [CrossRef]
- Ni, Z.; Cao, Y.; Jin, X.; Fu, Z.; Li, J.; Mo, X.; He, Y.; Tang, J.; Huang, S. Engineering resistance to bacterial blight and bacterial leaf streak in rice. Rice 2021, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Abdelrahman, M.; Gao, Y.; Ji, Z.; Mishra, R.; Sun, H.; Sui, Y.; Wu, C.; Wang, C.; Zhao, K. Engineering broad-spectrum resistance to bacterial blight by CRISPR-Cas9-mediated precise homology directed repair in rice. Mol. Plant 2021, 14, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, S.; Li, H.; Song, C.; Xie, W.; Zuo, S.; Zhou, X.; Zhou, C.; Ji, Z.; Zhou, H. Genome editing of a dominant resistance gene for broad-spectrum resistance to bacterial diseases in rice without growth penalty. Plant Biotechnol. J. 2024, 22, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, M.; Wang, Z.; Fang, R.; Wang, X.; Jia, Y. Dynamic and coordinated expression changes of rice small RNAs in response to Xanthomonas oryzae pv. oryzae. J. Genet. Genom. 2015, 42, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Liu, Y.; Zhang, H.; Xiao, J.; Li, X.; Wang, S. Small RNAs and gene network in a durable disease resistance gene--mediated defense responses in rice. PLoS ONE 2015, 10, e0137360. [Google Scholar] [CrossRef]
- Lu, J.; Wang, C.; Zhang, F.; Zeng, D.; Zhou, Y. Comparative microRNA profiling reveals microRNAs involved in rice resistant response to bacterial blight. Crop J. 2021, 9, 834–842. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar]
- Yuan, N.; Yuan, S.; Li, Z.; Li, D.; Hu, Q.; Luo, H. Heterologous expression of a rice miR395 gene in Nicotiana tabacum impairs sulfate homeostasis. Sci. Rep. 2016, 6, 28791. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Oono, Y.; Zhu, J.; He, X.; Wu, J.; Iida, K.; Lu, X.; Cui, X.; Jin, H.; Zhu, J. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ding, H.; Zhu, J.; Zhang, F.; Li, W. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, L.; Li, W.; Hu, X.; Wang, M.; Fan, Y.; Zhang, C.; Wang, L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhu, M.; Luan, M.; Zhang, M.; Jin, L.; Liu, Y.; Zou, J.; Wang, L.; Xu, M. miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 2022, 188, 608–623. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Combier, J.P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernie, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef]
- Zhao, B.; Ge, L.; Liang, R.; Li, W.; Ruan, K.; Lin, H.; Jin, Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009, 10, 29. [Google Scholar] [CrossRef]
- Liu, J.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Addo-Quaye, C.; Zhang, L.; Saini, A.; Jagadeeswaran, G.; Axtell, M.J.; Zhang, W.; Sunkar, R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010, 62, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Gu, L.; Li, P.; Song, X.; Wei, L.; Chen, Z.; Cao, X. Degradome sequencing reveals endogenous small RNAtargets in rice (Oryza sativa L. ssp. indica). Front. Biol. 2010, 5, 67–90. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef]
- Gautam, K.; Rao, P.B.; Chauhan, S.V.S. Efficacy of some botanicals of the family compositae against Rhizoctonia solani Kuhn. J. Mycol. Plant Pathol. 2003, 33, 230–235. [Google Scholar]
- Rao, T.B.; Chopperla, R.; Prathi, N.B.; Balakrishnan, M.; Prakasam, V.; Laha, G.S.; Balachandran, S.M.; Mangrauthia, S.K. A comprehensive gene expression profile of pectin degradation enzymes reveals the molecular events during cell wall degradation and pathogenesis of rice sheath blight pathogen Rhizoctonia solani AG1-IA. J. Fungi 2020, 6, 71. [Google Scholar] [CrossRef]
- Zou, J.; Pan, X.; Chen, Z.; Xu, J.; Lu, J.; Zhai, W.; Zhu, L. Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor. Appl. Genet. 2000, 101, 569–573. [Google Scholar] [CrossRef]
- Wang, A.; Shu, X.; Jing, X.; Jiao, C.; Chen, L.; Zhang, J.; Ma, L.; Jiang, Y.; Yamamoto, N.; Li, S.; et al. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 2021, 19, 1553–1566. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Karabi Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef]
- Chopperla, R.; Mangrauthia, S.K.; Rao, T.B.; Balakrishnan, M.; Balachandran, S.M.; Prakasam, V.; Channappa, G. A comprehensive analysis of microRNAs expressed in susceptible and resistant rice cultivars during Rhizoctonia solani AG1-IA infection causing sheath blight disease. Int. J. Mol. Sci. 2020, 21, 7974. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; He, L.; He, J.; Qin, P.; Wang, Y.; Deng, Q.; Yang, X.; Li, S.; Wang, S.; Wang, W.; et al. Comprehensive analysis of microRNA-Seq and target mRNAs of rice sheath blight pathogen provides new insights into pathogenic regulatory mechanisms. DNA Res. 2016, 23, 415–425. [Google Scholar] [CrossRef]
- Cao, W.; Cao, X.; Zhao, J.; Zhang, Z.; Feng, Z.; OuYang, S.; Zuo, S. Comprehensive characteristics of microRNA expression profile conferring to Rhizoctonia solani in rice. Rice Sci. 2020, 27, 101–112. [Google Scholar]
- Sasani, S.T.; Soltani, B.M.; Mehrabi, R.; Padasht-Dehkaei, H.S. Expression alteration of candidate rice miRNAs in response to sheath blight disease. Iran. J. Biotechnol. 2020, 18, e2451. [Google Scholar]
- Huang, J.; Li, Z.; Zhao, D. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 2016, 6, 29938. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Liu, Y.; Chen, X. Exploring heat-response mechanisms of microRNAs based on microarray data of rice post-meiosis panicle. Int. J. Genom. 2020, 2020, 7582612. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, W.; Wen, K.; Chen, G.; Sun, J.; Tian, Y.; Tang, W.; Yu, J.; An, H.; Wu, T.; et al. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 2017, 95, 345–357. [Google Scholar] [CrossRef]
- Okuda, K.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Shikanai, T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 2007, 104, 8178–8183. [Google Scholar] [CrossRef]
- Okuda, K.; Chateigner-Boutin, A.-L.; Nakamura, T.; Delannoy, E.; Sugita, M.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Small, I.; Shikanai, T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA Editing and RNA Cleavage in Arabidopsis chloroplasts. Plant Cell 2009, 21, 146–156. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef]
- Boussardon, C.; Salone, V.; Avon, A.; Berthome, R.; Hammani, K.; Okuda, K.; Shikanai, T.; Small, I.; Lurin, C. Two interacting proteins are necessary for the editing of the NdhD-1 Site in Arabidopsis plastids. Plant Cell 2012, 24, 3684–3694. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Francs-Small, C.; Fujii, S.; Okuda, K.; Tanz, S.K.; Small, I. The E domains of pentatricopeptide repeat proteins from different organelles are not functionally equivalent for RNA editing. Plant J. 2013, 74, 935–945. [Google Scholar] [CrossRef]
- Hayes, M.L.; Giang, K.; Berhane, B.; Mulligan, R.M. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-Terminal cytidine deaminase-like Domains. J. Biol. Chem. 2013, 288, 36519–36529. [Google Scholar] [CrossRef]
- Schallenberg-Rudinger, M.; Kindgren, P.; Zehrmann, A.; Small, I.; Knoop, V. A DYW-protein knockout in Physcomitrella affects two closely spaced mitochondrial editing sites and causes a severe developmental phenotype. Plant J. 2013, 76, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Wagoner, J.A.; Sun, T.; Lin, L.; Hanson, M.R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA Editing. J. Biol. Chem. 2015, 290, 2957–2968. [Google Scholar] [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis mitochondrialocalized pentatricopeptide repeat rrotein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011, 156, 2053–2068. [Google Scholar] [CrossRef]
- Jiao, X.; Wang, H.; Yan, J.; Kong, X.; Liu, Y.; Chu, J.; Chen, X.; Fang, R.; Yan, Y. Promotion of BR biosynthesis by miR444 is required for ammonium triggered inhibition of root growth. Plant Physiol. 2020, 182, 1454–1466. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. MiR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef]
- Zhou, R.; Sanz-Jimenez, P.; Zhu, X.; Feng, J.; Shao, L.; Song, J.; Chen, L. Analysis of rice transcriptome reveals the LncRNA/CircRNA regulation in tissue development. Rice 2021, 28, 14. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.; Thomma, B.; Huang, H.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]

| Pathogens | osa-miRNAs | Target Genes | Role | References |
|---|---|---|---|---|
| M. oryzae | osa-miR169 | OsNF-YA2 | Negative | [40] |
| osa-miR167d | OsARF12 | Negative | [41] | |
| osa-miR171b | OsSCL6-IIa, OsSCL6-IIb, OsSCL6-IIc | Positive | [42] | |
| osa-miR319 | OsTCP21 | Negative | [43] | |
| osa-miR535 | OsSPL4 | Negative | [44] | |
| osa-miR160a | OsARF8 | Positive | [45,46] | |
| osa-miR164a | OsNAC60 | Negative | [47] | |
| osa-miR156fhl | OsSPL14 | Negative | [48] | |
| osa-miR398b | OsCSD1, OsCSD2, OsSODX, OsCCSD | Positive | [46] [49] | |
| osa-miR7695 | OsNramp6 | Positive | [50] [51] [52] | |
| osa-miR166h-166k | OsEIN2 | Positive | [53] | |
| osa-miR396 | OsGRF6, OsGRF7, OsGRF8, OsGRF9 | Negative | [54] | |
| osa-miR162a | OsDCL1 | Positive | [55] | |
| osa-miR168 | OsAGO1 | Negative | [56] | |
| osa-miR1875 | OsHXK1 | Negative | [57] | |
| osa-miR812w | OsACO3, OsCIPK10, OsLRR | Positive | [58] | |
| osa-miR1873 | LOC_Os05g01790 | Negative | [59] | |
| osa-miR159a | OsGAMYB, OsGAMYBL, OsZF | Positive | [60] | |
| osa-miR1871 | OsMFAP1 | Negative | [61] | |
| osa-miR439 | LOC_Os01g23940, LOC_Os01g36270 | Negative | [62] | |
| osa-miR530 | OsDHFR-TS | Negative | [63] | |
| osa-miR1432 | OsEFH1, OsACOT | Negative | [64] | |
| Xoo | osa-miR160a | OsARF8 | Positive | [45] |
| osa-miR156 | OsSPL14 (IPA1), OsSPL7 | Negative | [65] | |
| osa-miRNA156/529 | OsSPL7/14/17 | Negative | [66] | |
| osa-miR169o | OsNF-YA1, OsNF-YA4 | Negative | [67] | |
| osa-miR1876 | OsNBS8R OsNBS8S | Negative | [68] | |
| osa-miR159b | OsGAMYB | Positive | [69] | |
| osa-miR164a | OsNAC60 | Negative | [69] | |
| osa-miR167d | OsWD40-174 | Negative | [69] | |
| osa-miR1432 | OsCaML2 | Positive | [70] | |
| osa-miR395 | OsAPS1, OsSULTR2;1, OsSULTR2;2 | Positive | [71] | |
| osa-miR2118 | LOC_Os08g42700.1, LOC_Os01g05600.1, LOC_Os12g37290.1 | Negative | [72] | |
| R. solani | osa-miR160a | OsARF8 | Positive | [45] |
| osa-miR395a | Os12t0109300 | Negative | [73] | |
| osa-miR444.2 | - | Negative | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Wei, K.; Qin, J.; Zhai, W.; Li, Q.; Li, Y. The Roles of MicroRNAs in the Regulation of Rice–Pathogen Interactions. Plants 2025, 14, 136. https://doi.org/10.3390/plants14010136
Jia Y, Wei K, Qin J, Zhai W, Li Q, Li Y. The Roles of MicroRNAs in the Regulation of Rice–Pathogen Interactions. Plants. 2025; 14(1):136. https://doi.org/10.3390/plants14010136
Chicago/Turabian StyleJia, Yanfeng, Kai Wei, Jiawang Qin, Wenxue Zhai, Quanlin Li, and Yalan Li. 2025. "The Roles of MicroRNAs in the Regulation of Rice–Pathogen Interactions" Plants 14, no. 1: 136. https://doi.org/10.3390/plants14010136
APA StyleJia, Y., Wei, K., Qin, J., Zhai, W., Li, Q., & Li, Y. (2025). The Roles of MicroRNAs in the Regulation of Rice–Pathogen Interactions. Plants, 14(1), 136. https://doi.org/10.3390/plants14010136






