Spatial Distribution of Genetic, Ploidy, and Morphological Variation of the Edaphic Steno-Endemic Alyssum moellendorfianum (Brassicaceae) from the Western Balkans
Abstract
:1. Introduction
2. Results
2.1. Flow Cytometry Analysis
2.2. Microsatellite Analyses
2.3. AFLP Fingerprinting
2.4. Chloroplast DNA Analysis
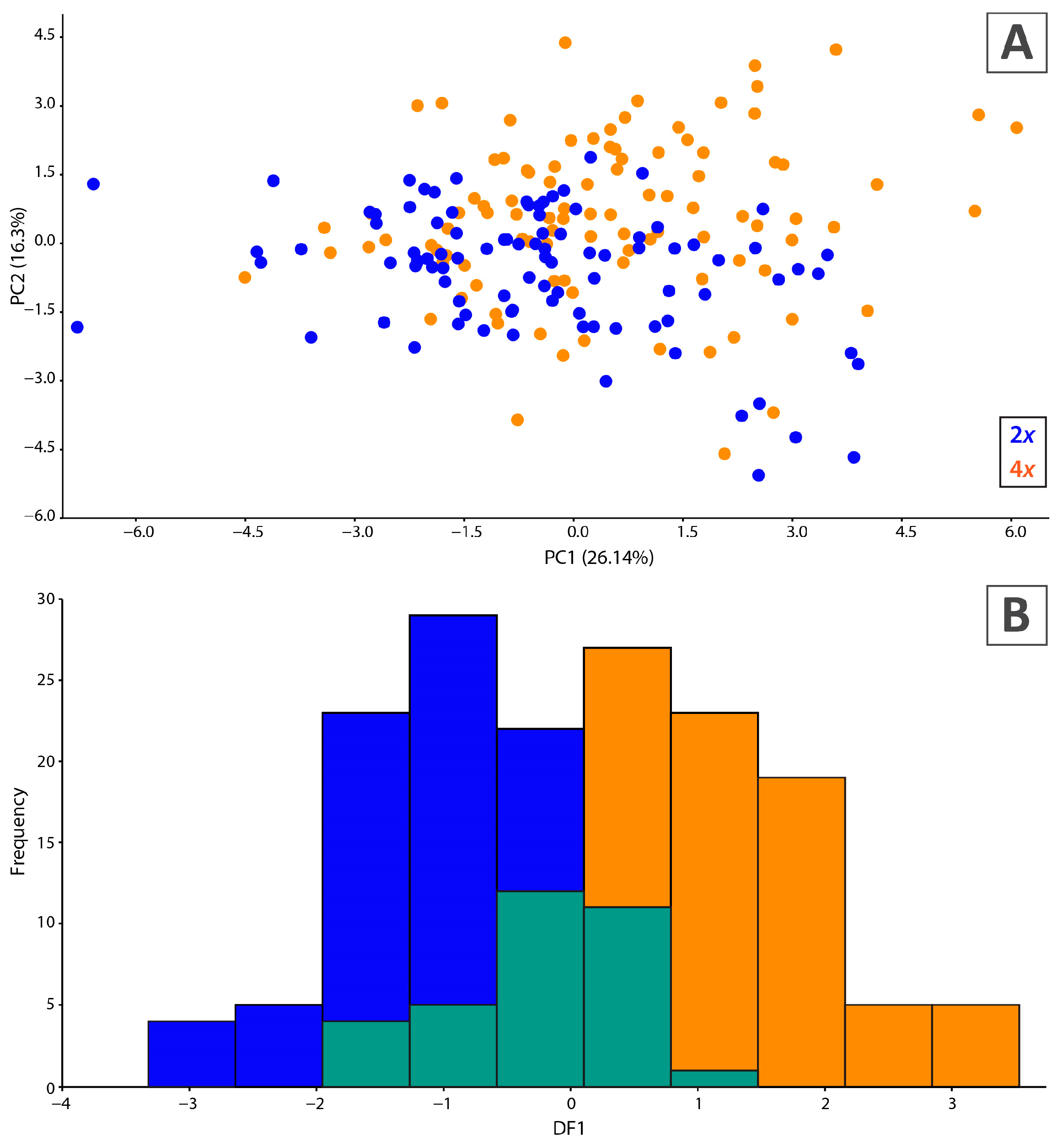
2.5. Morphological Variation of Cytotypes
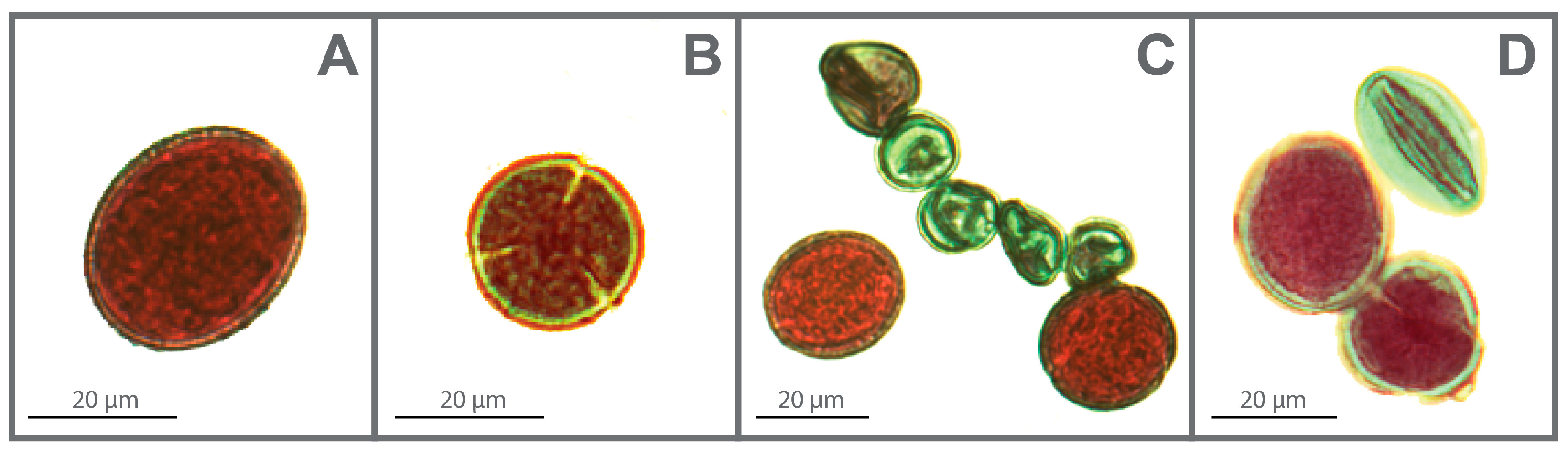
2.6. Size and Viability of Pollen Grains
3. Discussion
3.1. The Spatial Distribution of Cytotypes
3.2. Genetic Diversity and Structure of Alyssum moellendorfianum
3.3. From Diploids to Tetraploids: Challenges in Polyploid Origin Reconstruction
3.4. Implications for Conservation
4. Materials and Methods
4.1. Plant Material
4.2. Genome Size and Ploidy Level Determination
4.3. DNA Extraction and Molecular Analysis
4.4. Microsatellite Genotyping
4.5. AFLP Fingerprinting
4.6. Chloroplast DNA Analyses: rpoB-trnC and rpl32-trnLUAG Intergenic Spacers
4.7. Morphometric Analyses
4.8. Pollen Grain Measurements and Viability Estimation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kryštufek, B.; Reed, J.M. Pattern and process in Balkan biodiversity—An overview. In Balkan Biodiversity; Griffiths, H.I., Kryštufek, B., Reed, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 1–8. [Google Scholar] [CrossRef]
- Stevanović, V.; Tan, K.; Petrova, A. Mapping the endemic flora of the Balkans—A progress report. Bocconea 2007, 21, 131–137. [Google Scholar]
- Hewitt, G.M. Mediterranean peninsulas: The evolution of hotspots. In Biodiversity Hotspots. Distribution and Protection of Conservation; Frank, E., Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin, Germany, 2011; pp. 123–147. [Google Scholar] [CrossRef]
- Nieto Feliner, G. Patterns and processes in plant phylogeography in the Mediterranean Basin. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 265–278. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean, 2nd ed.; Oxford University Press: Oxford, UK, 2020. [Google Scholar] [CrossRef]
- López-González, N.; Bobo-Pinilla, J.; Padilla-García, N.; Loureiro, J.; Castro, S.; Rojas-Andrés, B.M.; Martínez-Ortega, M.M. Genetic similarities versus morphological resemblance: Unraveling a polyploid complex in a Mediterranean biodiversity hotspot. Mol. Phylogenet Evol. 2021, 155, 107006. [Google Scholar] [CrossRef]
- Šlenker, M.; Kantor, A.; Marhold, K.; Schmickl, R.; Mandáková, T.; Lysak, M.A.; Perný, M.; Caboňová, M.; Slovák, M.; Zozomová-Lihová, J. Allele sorting as a novel approach to resolving the origin of allotetraploids using Hyb-Seq data: A case study of the Balkan mountain endemic Cardamine barbaraeoides. Front. Plant Sci. 2021, 12, 659275. [Google Scholar] [CrossRef] [PubMed]
- Bognar, A.; Faivre, S.; Pavelić, J. Glaciation traces on the Northern Velebit. Geogr. Glas. 1991, 53, 27–39. [Google Scholar]
- Milivojević, M.; Menković, L.; Ćalić, J. Pleistocene glacial relief of the central part of Mt. Prokletije (Albanian Alps). Quat. Int. 2008, 190, 112–122. [Google Scholar] [CrossRef]
- Gömöry, D.; Zhelev, P.; Brus, R. The Balkans: A genetic hotspot but not a universal colonization source for trees. Plant Syst. Evol. 2020, 306, 5. [Google Scholar] [CrossRef]
- Rešetnik, I.; Záveská, E.; Grgurev, M.; Bogdanović, S.; Bartolić, P.; Frajman, B. Stability in the South, Turbulence Toward the North: Evolutionary History of Aurinia saxatilis (Brassicaceae) Revealed by Phylogenomic and Climatic Modelling Data. Front. Plant Sci. 2022, 13, 822331. [Google Scholar] [CrossRef] [PubMed]
- Španiel, S.; Rešetnik, I. Plant phylogeography of the Balkan Peninsula: Spatiotemporal patterns and processes. Plant Syst. Evol. 2022, 308, 38. [Google Scholar] [CrossRef]
- Kutnjak, D.; Kuttner, M.; Niketić, M.; Dullinger, S.; Schönswetter, P.; Frajman, B. Escaping to the summits: Phylogeography and predicted range dynamics of Cerastium dinaricum, an endangered high mountain plant endemic to the western Balkan Peninsula. Mol. Phylogenet Evol. 2014, 78, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Đurović, S.Z.; Temunović, M.; Niketić, M.; Tomović, G.; Schönswetter, P.; Frajman, B. Impact of Quaternary climatic oscillations on phylogeographic patterns of three habitat-segregated Cerastium taxa endemic to the Dinaric Alps. J. Biogeogr. 2021, 48, 2022–2036. [Google Scholar] [CrossRef]
- Tomović, G.; Niketić, M.; Lakušić, D.; Ranđelović, V.; Stevanović, V. Balkan endemic plants in Central Serbia and Kosovo regions: Distribution patterns, ecological characteristics, and centres of diversity. Bot. J. Linn. Soc. 2014, 176, 173–202. [Google Scholar] [CrossRef]
- Jug-Dujaković, M.; Ninčević, T.; Liber, Z.; Grdiša, M.; Šatović, Z. Salvia officinalis survived in situ Pleistocene glaciation in ‘refugia within refugia’ as inferred from AFLP markers. Plant Syst. Evol. 2020, 306, 38. [Google Scholar] [CrossRef]
- Caković, D.; Stešević, D.; Schönswetter, P.; Frajman, B. Long neglected diversity in the Accursed Mountains of northern Albania: Cerastium hekuravense is genetically and morphologically divergent from C. dinaricum. Plant Syst. Evol. 2018, 304, 57–69. [Google Scholar] [CrossRef]
- Liber, Z.; Surina, B.; Nikolić, T.; Škrtić, D.; Šatović, Z. Spatial distribution, niche ecology and conservation genetics of Degenia velebitica (Brassicaceae), a narrow endemic species of the north-western Dinaric Alps. Plant Syst. Evol. 2020, 306, 64. [Google Scholar] [CrossRef]
- Ninčević, T.; Jug-Dujaković, M.; Grdiša, M.; Liber, Z.; Varga, F.; Pljevljakušić, D.; Šatović, Z. Population structure and adaptive variation of Helichrysum italicum (Roth) G. Don along eastern Adriatic temperature and precipitation gradient. Sci. Rep. 2021, 11, 24333. [Google Scholar] [CrossRef]
- Zozomová-Lihová, J.; Melichárková, A.; Svitok, M.; Španiel, S. Pleistocene range disruption and postglacial expansion with secondary contacts explain the genetic and cytotype structure in the western Balkan endemic Alyssum austrodalmaticum (Brassicaceae). Plant Syst. Evol. 2020, 306, 47. [Google Scholar] [CrossRef]
- Radosavljević, I.; Satovic, Z.; di Pietro, R.; Jug Dujaković, M.; Varga, F.; Škrtić, D.; Liber, Z. Phylogeographic structure of common sage (Salvia ofcinalis L.) reveals microrefugia throughout the Balkans and colonizations of the Apennines. Sci. Rep. 2022, 12, 15726. [Google Scholar] [CrossRef]
- Hajrudinović, A.; Frajman, B.; Schönswetter, P.; Silajdžić, E.; Siljak-Yakovlev, S.; Bogunić, F. Towards a better understanding of polyploid Sorbus (Rosaceae) from Bosnia and Herzegovina (Balkan Peninsula), including description of a novel, tetraploid apomictic species. Bot. J. Linn. Soc. 2015, 178, 670–685. [Google Scholar] [CrossRef]
- Hajrudinovic, A.; Siljak-Yakovlev, S.; Brown, S.C.; Pustahija, F.; Bourge, M.; Ballian, D.; Bogunic, F. When sexual meets apomict—Genome size, ploidy level and reproduction mode variation of Sorbus aria s.l. and S. austriaca (Rosaceae) in Bosnia and Herzegovina. Ann. Bot. 2015, 116, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Lazarević, M.; Siljak-Yakovlev, S.; Sanino, A.; Niketić, M.; Lamy, F.; Hinsinger, D.D.; Tomović, G.; Stevanović, B.; Stevanović, V.; Robert, T. Genetic variability in Balkan paleoendemic resurrection plants Ramonda serbica and R. nathaliae across their range and in the zone of sympatry. Front. Plant Sci. 2022, 13, 873471. [Google Scholar] [CrossRef]
- Kuzmanović, N.; Comanescu, P.; Frajman, B.; Lazarević, M.; Paun, O.; Schönswetter, P.; Lakušić, D. Genetic, cytological and morphological differentiation within the Balkan-Carpathian Sesleria rigida sensu Fl. Eur. (Poaceae): A taxonomically intricate tetraploid-octoploid complex. Taxon 2013, 62, 458–472. [Google Scholar] [CrossRef]
- Kuzmanović, N.; Lakušić, D.; Stevanoski, I.; Barfuss, M.H.J.; Schönswetter, P.; Frajman, B. Carpinus austrobalcanica—A new highly polyploid species from the Balkan Peninsula closely related to European hornbeam. Perspect. Plant Ecol. Evol. Syst. 2024, 64, 125812. [Google Scholar] [CrossRef]
- Rešetnik, I.; Schonswetter, P.; Temunović, M.; Barfuss, M.H.J.; Frajman, B. Diploid chastity vs. polyploid promiscuity—Extensive gene flow among polyploid cytotypes blurs genetic, morphological and taxonomic boundaries among Dinaric taxa of Knautia (Caprifoliaceae). Perspect. Plant Ecol. Evol. Syst. 2023, 59, 125730. [Google Scholar] [CrossRef]
- Niketić, M.; Đurović, S.Z.; Tomović, G.; Schönswetter, P.; Frajman, B. Diversification within ploidy-variable Balkan endemic Cerastium decalvans (Caryophyllaceae) reconstructed based on genetic, morphological and ecological evidence. Bot. J. Linn. Soc. 2022, 199, 578–608. [Google Scholar] [CrossRef]
- Španiel, S.; Kempa, M.; Salmeron-Sanchez, E.; Fuertes-Aguilar, J.; Mota, J.F.; Al-Shehbaz, I.A.; German, D.A.; Olsavska, K.; Singliarova, B.; Zozomová-Lihova, J.; et al. AlyBase: Database of names, chromosome numbers, and ploidy levels of Alysseae (Brassicaceae), with a new generic concept of the tribe. Plant Syst. Evol. 2015, 301, 2463–2491. [Google Scholar] [CrossRef]
- Španiel, S.; Marhold, K.; Zozomova-Lihova, J. The polyploid Alyssum montanum-A. repens complex in the Balkans: A hotspot of species and genetic diversity. Plant Syst. Evol. 2017, 303, 1443–1465. [Google Scholar] [CrossRef]
- Španiel, S.; Zozomová-Lihová, J.; Marhold, K. Revised taxonomic treatment of the Alyssum montanum-A. repens complex in the Balkans: A multivariate morphometric analysis. Plant Syst. Evol. 2017, 303, 1413–1442. [Google Scholar] [CrossRef]
- Magauer, M.; Schonswetter, P.; Jang, T.S.; Frajman, B. Disentangling relationships within the disjunctly distributed Alyssum ovirense/A. wulfenianum group (Brassicaceae), including description of a novel species from the north-eastern Alps. Bot. J. Linn. Soc. 2014, 176, 486–505. [Google Scholar] [CrossRef]
- Zozomová-Lihová, J.; Marhold, K.; Španiel, S. Taxonomy and evolutionary history of Alyssum montanum (Brassicaceae) and related taxa in southwestern Europe and Morocco: Diversification driven by polyploidy, geographic and ecological isolation. Taxon 2014, 63, 562–591. [Google Scholar] [CrossRef]
- Melichárková, M.; Španiel, S.; Briskova, D.; Marhold, K.; Zozomova-Lihova, J. Unravelling allopolyploid origins in the Alyssum montanum-A. repens species complex (Brassicaceae): Low-copy nuclear gene data complement plastid DNA sequences and AFLPs. Bot. J. Linn. Soc. 2017, 184, 485–502. [Google Scholar] [CrossRef]
- Melichárková, M.; Španiel, S.; Marhold, K.; Hurdu, B.I.; Drescher, A.; Zozomová-Lihová, J. Diversification and independent polyploid origins in the disjunct species Alyssum repens from the Southeastern Alps and the Carpathians. Am. J. Bot. 2019, 106, 1499–1518. [Google Scholar] [CrossRef] [PubMed]
- Cetlová, V.; Zozomová-Lihová, J.; Melichárková, A.; Mártonfiová, L.; Španiel, S. Multiple drivers of high species diversity and endemism among Alyssum annuals in the Mediterranean: The evolutionary significance of the Aegean hotspot. Front. Plant Sci. 2021, 12, 627909. [Google Scholar] [CrossRef]
- Španiel, S.; Šlenker, M.; Melichárková, A.; Caboňová, M.; Šandalová, M.; Zeisek, V.; Marhold, K.; Zozomová-Lihová, J. Phylogenetic challenges in a recently diversified and polyploid-rich Alyssum (Brassicaceae) lineage: Low divergence, reticulation and parallel polyploid speciation. Evolution 2023, 77, 1226–1244. [Google Scholar] [CrossRef] [PubMed]
- Riter-Studnička, H. Flora i vegetacija na dolomitima Bosne i Hercegovine. Godišnjak Biološkog Instituta 1956, IX, 73–116. [Google Scholar]
- Red List of Endangered Wild Species and Subspecies of Plants, Animals and Fungi. Official Gazette of FBiH, 2014. no 7/14. Available online: https://faolex.fao.org/docs/pdf/bih204210.pdf (accessed on 9 November 2024).
- Siljak-Yakovlev, S.; Muratović, E.; Bogunić, F.; Bašić, N.; Šolić, M.; Boškailo, A.; Bourge, M.; Valentin, N.; Stevanović, V.; Garnatje, T.; et al. Genome size of Balkan flora: A database (GeSDaBaF) and C-values for 51 taxa of which 46 are novel. Plant Syst. Evol. 2020, 306, 40. [Google Scholar] [CrossRef]
- Bourge, M.; Brown, S.; Siljak-Yakovlev, S. Flow cytometry as tool in plant sciences, with emphasis on genome size and ploidy level assessment. Genet. Appl. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Soltis, D.; Buggs, R.; Doyle, J.; Soltis, P. What we still don’t know about polyploidy. Taxon 2010, 59, 1387–1403. [Google Scholar] [CrossRef]
- Trávníček, P.; Kubátová, B.; Čurn, V.; Rauchová, J.; Krajníková, E.; Jersáková, J.; Suda, J. Remarkable coexistence of multiple cytotypes of the Gymnadenia conopsea aggregate (the fragrant orchid): Evidence from flow cytometry. Ann. Bot. 2011, 107, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Castro, S.; Loureiro, J.; Procházka, T.; Münzbergová, Z. Cytotype distribution at a diploid-hexaploid contact zone in Aster amellus (Asteraceae). Ann. Bot. 2012, 110, 1047–1055. [Google Scholar] [CrossRef]
- Kolář, F.; Lučanová, M.; Vít, P.; Urfus, T.; Chrtek, J.; Fér, T.; Ehrendorfer, F.; Suda, J. Diversity and endemism in deglaciated areas: Ploidy, relative genome size and niche differentiation in the Galium pusillum complex (Rubiaceae) in Northern and Central Europe. Ann. Bot. 2013, 111, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Kubátová, B.; Trávníček, P.; Bastlová, D.; Čurn, V.; Jarolímová, V.; Suda, J. DNA ploidy-level variation in native and invasive populations of Lythrum salicaria at a large geographical scale. J. Biogeogr. 2008, 35, 167–176. [Google Scholar] [CrossRef]
- Castro, M.; Loureiro, J.; Husband, B.C.; Castro, S. The role of multiple reproductive barriers: Strong post-pollination interactions govern cytotype isolation in a tetraploid-octoploid contact zone. Ann. Bot. 2020, 126, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Čertner, M. Why is reproductive isolation between intraspecific cytotypes so important? A commentary on: “Strong post-pollination interactions govern cytotype isolation in a tetraploid-octoploid contact zone”. Ann. Bot. 2020, 126, iv–v. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.L.; Galloway, L.F. Postzygotic isolation varies by ploidy level within a polyploid complex. New Phytol. 2017, 213, 404–412. [Google Scholar] [CrossRef]
- Köhler, C.; Mittelsten Scheid, O.; Erilova, A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010, 26, 142–148. [Google Scholar] [CrossRef]
- Lakušić, D.; Liber, Z.; Nikolić, T.; Surina, B.; Kovačić, S.; Bogdanović, S.; Stefanović, S. Molecular phylogeny of the Campanula pyramidalis species complex (Campanulaceae) inferred from chloroplast and nuclear non-coding sequences and its taxonomic implications. Taxon 2013, 62, 505–524. [Google Scholar] [CrossRef]
- Glasnović, P.; Temunović, M.; Lakušić, D.; Rakić, T.; Brečko Grubar, V.; Surina, B. Understanding biogeographical patterns in the western Balkan Peninsula using environmental niche modelling and geostatistics in polymorphic Edraianthus tenuifolius. AoB Plants 2018, 10, ply064. [Google Scholar] [CrossRef] [PubMed]
- Rešetnik, I.; Baričevič, D.; Batîr Rusu, D.; Carović-Stanko, K.; Chatzopoulou, P.; Dajić-Stevanović, Z.; Gonceariuc, M.; Grdiša, M.; Greguraš, D.; Ibraliu, A.; et al. Genetic diversity and demographic history of wild and cultivated/naturalised plant populations: Evidence from Dalmatian Sage (Salvia officinalis L., Lamiaceae). PLoS ONE 2016, 11, e0159545. [Google Scholar] [CrossRef]
- Horvat, I.; Glavač, V.; Ellenberg, H. Vegetation Südosteuropas, 1st ed.; Gustav Fischer Verlag: Stuttgart, Germany, 1974. [Google Scholar]
- Roglić, J. Zaravni na vapnencima. Geogr. Glas. 1957, 19, 103–134. [Google Scholar]
- Roglić, J. Prilog poznavanju glacijacije i evolucije reljefa planina oko srednje Neretve. Geogr. Glas. 1959, 21, 9–34. [Google Scholar]
- Tokić, S. Genetski tipovi i osnovne geološke odlike kvartarnih naslaga Bosne i Hercegovine. Geološki Glas. 1983, 28, 219–252. [Google Scholar]
- Hrvatović, H. Geološki Vodič Kroz Bosnu i Hercegovinu, 1st ed.; Zavod za Geologiju: Sarajevo, Bosnia and Herzegovina, 1999. [Google Scholar]
- Çiner, A.; Stepišnik, U.; Sarıkaya, M.A.; Žebre, M.; Yıldırım, C. Last Glacial Maximum and Younger Dryas piedmont glaciations in Blidinje, the Dinaric Mountains (Bosnia and Herzegovina); insights from 36Cl cosmogenic dating. Mediterr. Geosci. Rev. 2019, 1, 25–43. [Google Scholar] [CrossRef]
- Žebre, M.; Sarıkaya, M.A.; Stepišnik, U.; Çiner, A. First 36Cl cosmogenic moraine geochronology of the Dinaric mountain karst: Velež and Crvanj Mountains of Bosnia and Herzegovina. Quat. Sci. Rev. 2019, 208, 54–75. [Google Scholar] [CrossRef]
- Riter-Studnička, H. Reliktgesellschaften auf Dolomitboden in Bosnien und der Hercegovina. Vegetatio 1967, 15, 190–212. [Google Scholar] [CrossRef]
- Gerth, A.; Merten, D.; Baumbach, H. Verbreitung, Vergesellschaftung und genetische Populationsdifferenzierung des Berg-Steinkrautes (Alyssum montanum L.) auf Schwermetallstandorten im östlichen Harzvorland. Hercynia 2011, 44, 73–92. [Google Scholar]
- Pojskić, N. ALRATIO-R script for the analysis of relation between the effective and the detected number of alleles. Genet. Appl. 2019, 3, 77–80. [Google Scholar] [CrossRef]
- Pojskić, N. iMAF—Index of Major Allele Frequency. Genet. Appl. 2018, 2, 78–81. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Population fitness is correlated with genetic diversity. Conserv. Biol. 2003, 7, 230–237. [Google Scholar] [CrossRef]
- Greenbaum, G.; Templeton, A.R.; Zarmi, Y.; Bar-David, S. Allelic richness following population founding events—A stochastic modeling framework incorporating gene flow and genetic drift. PLoS ONE 2014, 9, e115203. [Google Scholar] [CrossRef] [PubMed]
- Caballero, A.; Rodriguez-Ramilo, S.; Avila, V.; Fernandez, J. Management of genetic diversity of subdivided populations in conservation programmes. Conserv. Genet. 2010, 11, 409–419. [Google Scholar] [CrossRef]
- Wagner, A. Robustness and evolvability: A paradox resolved. Proc. Biol. Sci. 2008, 275, 91–100. [Google Scholar] [CrossRef] [PubMed]
- González, A.V.; Gómez-Silva, V.; Ramírez, M.J.; Fontúrbel, F.E. Meta-analysis of the differential effects of habitat fragmentation and degradation on plant genetic diversity. Conserv. Biol. 2020, 34, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, L.C.; Reis, M.N.O.; Bessa, L.A.; Souza, U.J.B.; Silva, F.G. Landscape and climate influence the patterns of genetic diversity and inbreeding in Cerrado plant species. Diversity 2020, 12, 421. [Google Scholar] [CrossRef]
- Španiel, S.; Marhold, K.; Filová, B.; Zozomová-Lihová, J. Genetic and morphological variation in the diploid-polyploid Alyssum montanum in Central Europe: Taxonomic and evolutionary considerations. Plant Syst. Evol. 2011, 294, 1–25. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci. 1993, 12, 243–273. [Google Scholar] [CrossRef]
- Kolář, F.; Čertner, M.; Suda, J.; Schönswetter, P.; Husband, B.C. Mixed-ployidy species: Progress and opportunities in polyploid research. Trends Plant Sci. 2017, 22, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, M.J.; Hiscock, S.J. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 2008, 18, 435–444. [Google Scholar] [CrossRef]
- Laport, R.G.; Ng, J. Out of one, many: The biodiversity considerations of polyploidy. Am. J. Bot. 2017, 104, 1119–1121. [Google Scholar] [CrossRef] [PubMed]
- Balao, F.; Herrera, J.; Talavera, S. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: A multivariate morphological approach. New Phytol. 2011, 192, 256–265. [Google Scholar] [CrossRef]
- Durkovič, J.; Kardošová, M.; Čaňová, I.; Lagaňa, R.; Priwitzer, T.; Chorvát, D.; Cicák, A.; Pichler, V. Leaf traits in parental and hybrid species of Sorbus (Rosaceae). Am. J. Bot. 2012, 99, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.A.; Husband, B.C.; Maherali, H. Climatic niche differences between diploid and tetraploid cytotypes of Chamerion angustifolium (Onagraceae). Am. J. Bot. 2014, 101, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Hörandl, E. The complex causality of geographical parthenogenesis. New Phytol. 2006, 171, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Soltis, P.S.; Schemske, D.W.; Hancock, J.F.; Thompson, J.N.; Husband, B.C.; Judd, W.S. Autopolyploidy in angiosperms: Have we grossly underestimated the number of species? Taxon 2007, 56, 3–30. [Google Scholar] [CrossRef]
- Jorgensen, M.H.; Carlsen, T.; Skrede, I.; Elven, R. Microsatellites resolve the taxonomy of the polyploid Cardamine digitata aggregate (Brassicaceae). Taxon 2008, 57, 882–892. [Google Scholar] [CrossRef]
- Feulner, M.; Weig, A.; Paule, J.; Gregor, T.; Schott, L.F.; Aas, G. Genetic variability and morphology of tri- and tetraploid members of the Sorbus aria complex in northern Bavaria. Preslia 2017, 89, 275–290. [Google Scholar] [CrossRef]
- Bigl, K.; Paule, J.; Dobeš, C. The morphometrics of autopolyploidy: Insignificant differentiation among sexual–apomictic cytotypes. AoB Plants 2019, 11, plz028. [Google Scholar] [CrossRef] [PubMed]
- Pungaršek, Š.; Frajman, B. Influence of polyploidy on morphology and distribution of the Cypress Spurge (Euphorbia cyparissias, Euphorbiaceae). Plant Biol. J. 2024, 26, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.M.; Forni-Martins, E.R.; Magalhães, P.M.; Alves, M.N. Chromosomal and morphological studies of diploid and polyploid cytotypes of Stevia rebaudiana (Bertoni) Bertoni (Eupatorieae, Asteraceae). Genet. Mol. Biol. 2004, 27, 215–222. [Google Scholar] [CrossRef]
- Anamthawat-Jónsson, K.; Thórsson, Æ.T.; Temsch, E.M.; Greilhuber, J. Icelandic birch polyploids—The case of a perfect fit in genome size. J. Bot. 2010, 2010, 347254. [Google Scholar] [CrossRef]
- Lazarević, M.; Siljak-Yakovlev, S.; Lazarević, P.; Stevanović, P.; Stevanović, V. Pollen and seed morphology of resurrection plants from the genus Ramonda (Gesneriaceae): Relationship with ploidy level and ecology. Turk. J. Bot. 2013, 37, 872–885. [Google Scholar] [CrossRef]
- Marinho, R.C.; Mendes-Rodrigues, C.; Bonetti, A.M.; Oliveira, P.E. Pollen and stomata morphometrics and polyploidy in Eriotheca (Malvaceae-Bombacoideae). Plant Biol. 2014, 16, 508–511. [Google Scholar] [CrossRef]
- Rotreklová, O.; Krahulcová, A. Estimating paternal efficiency in an agamic polyploid complex: Pollen stainability and variation in pollen size related to reproduction mode, ploidy level, and hybridogenous origin in Pilosella (Asteraceae). Folia Geobot. 2016, 51, 175–186. [Google Scholar] [CrossRef]
- Schinkel, C.C.F.; Kirchheimer, B.; Dullinger, S.; Geelen, D.; Storme, N.D.; Hörandl, E. Pathways to polyploidy: Indications of a female triploid bridge in the alpine species Ranunculus kuepferi (Ranunculaceae). Plant Syst. Evol. 2017, 303, 1093–1108. [Google Scholar] [CrossRef]
- Brochmann, C. Pollen and seed morphology of Nordic Draba (Brassicaceae): Phylogenetic and ecological implications. Nord. J. Bot. 1992, 12, 657–673. [Google Scholar] [CrossRef]
- Baser, B.; Baygledi, Z.; Sahin, B. Pollen morphology of some Alyssum, L. (Brassicaceae) from Turkey. J. Sci. Technol. 2018, 8, 60–68. [Google Scholar]
- Al-Blesh, M.A.H.; Al-Joaany, M.D.S. The palynological study of ten species of Alyssum (Brassicaceae) in Mid and North Iraq. Plant Arch. 2020, 20, 413–416. [Google Scholar]
- Erden, A.; Menemen, Y. Comparative pollen morphology studies on some species of Brassicaceae in Turkey. Biol. Divers. Conserv. 2021, 14, 105–118. [Google Scholar] [CrossRef]
- Luque, T.; Díaz Lifante, Z. Chromosome numbers of plants collected during Iter Mediterraneum I in the SE of Spain. Bocconea 1991, 1, 303–364. [Google Scholar]
- Ghaffari, S.M.; Chariat-Panahi, M.S. Chromosome counts of some Angiosperms from Iran. Iran. J. Bot. 1985, 3, 68–73. [Google Scholar]
- Díaz Lifante, Z.; Parra Martín, R. Chromosome numbers of plants collected during Iter Mediterraneum V in Morocco. Bocconea 2013, 26, 151–172. [Google Scholar] [CrossRef]
- Kraus, D.; Enns, A.; Hebb, A.; Murphy, S.; Drake, D.A.R.; Bennett, B. Prioritizing nationally endemic species for conservation. Conserv. Sci. Pract. 2023, 5, e12845. [Google Scholar] [CrossRef]
- Cires, E.; Sanna, M.; Vázquez, V.M.; Prieto, J.A.F. Genetics and conservation of rare and endemic plants: The case of Genista sanabrensis (Fabaceae) in the Iberian Peninsula. Mediterr. Bot. 2018, 39, 77–87. [Google Scholar] [CrossRef]
- Winkler, D.E.; Lin, M.Y.; Delgadillo, J.; Chapin, K.J.; Huxman, T.E. Early life history responses and phenotypic shifts in a rare endemic plant responding to climate change. Conserv. Physiol. 2019, 7, coz076. [Google Scholar] [CrossRef] [PubMed]
- Hussain Mir, A.; Tyub, S.; Kamili, A.N. Ecology, distribution mapping and conservation implications of four critically endangered endemic plants of Kashmir Himalaya. Saudi J. Biol. Sci. 2020, 27, 2380–2389. [Google Scholar] [CrossRef] [PubMed]
- Kougioumoutzis, K.; Kokkoris, I.P.; Panitsa, M.; Strid, A.; Dimopoulos, P. Extinction Risk Assessment of the Greek Endemic Flora. Biology 2021, 10, 195. [Google Scholar] [CrossRef]
- Hvilsom, C.; Segelbacher, G.; Ekblom, R.; Fischer, M.C.; Laikre, L.; Leus, K.; O’Brien, D.; Shaw, R.; Sork, V. Selecting Species and Populations for Monitoring of Genetic Diversity; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2022. [Google Scholar] [CrossRef]
- Geppert, C.; Perazza, G.; Wilson, R.J.; Bertolli, A.; Prosser, F.; Melchiori, G.; Marini, L. Consistent population declines but idiosyncratic range shifts in Alpine orchids under global change. Nat. Commun. 2020, 11, 5835. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Q. Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant Biol. 2020, 20, 510. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Cullings, K.W. Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Mol. Ecol. 1992, 1, 233–240. [Google Scholar] [CrossRef]
- Brody, J.R.; Kern, S.E. Sodium boric acid: A Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques 2004, 36, 214–216. [Google Scholar] [CrossRef]
- Sobczyk, M.K.; Smith, J.A.C.; Pollard, A.J.; Filatov, D.A. Evolution of nickel hyperaccumulation and serpentine adaptation in the Alyssum serpyllifolium species complex. Heredity 2017, 118, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.V.; Jasieniuk, M. POLYSAT: An R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Trybush, S.; Hanley, S.; Cho, K.H.; Jahodova, S.; Grimmer, M.; Emelianov, I.; Bayon, C.; Karp, A. Getting the most out of fluorescent amplified fragment length polymorphism. Can. J. Bot. 2006, 84, 1347–1354. [Google Scholar] [CrossRef]
- Bonin, A.; Bellemain, E.; Bronken Eidesen, P.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261–3273. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; De Wachter, R. TREECON: A software package for the construction and drawing of evolutionary trees. Bioinformatics 1997, 9, 177–182. [Google Scholar] [CrossRef]
- Bryant, D.; Moulton, V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Shaw, J.; Lickey, E.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2008, 36, D25–D30. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Librado, P.; Guirao-Rico, S.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2018, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Ft. Lauderdale, FL, USA, 15–19 April 2002; Volume 2, p. 184. [Google Scholar]
- Quinn, G.P.; Keough, M.J. Experimental Design and Data Analysis for Biologists; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Wodehouse, R.P. Pollen Grains. Their Structure, Identification and Significance in Science and Medicine; Hafner Publish Company: New York, NY, USA; London, UK, 1965; pp. 106–109. [Google Scholar]
- Alexander, M.P. Differential staining of aborted and non-aborted pollen. Stain. Technol. 1969, 44, 117–122. [Google Scholar] [CrossRef] [PubMed]
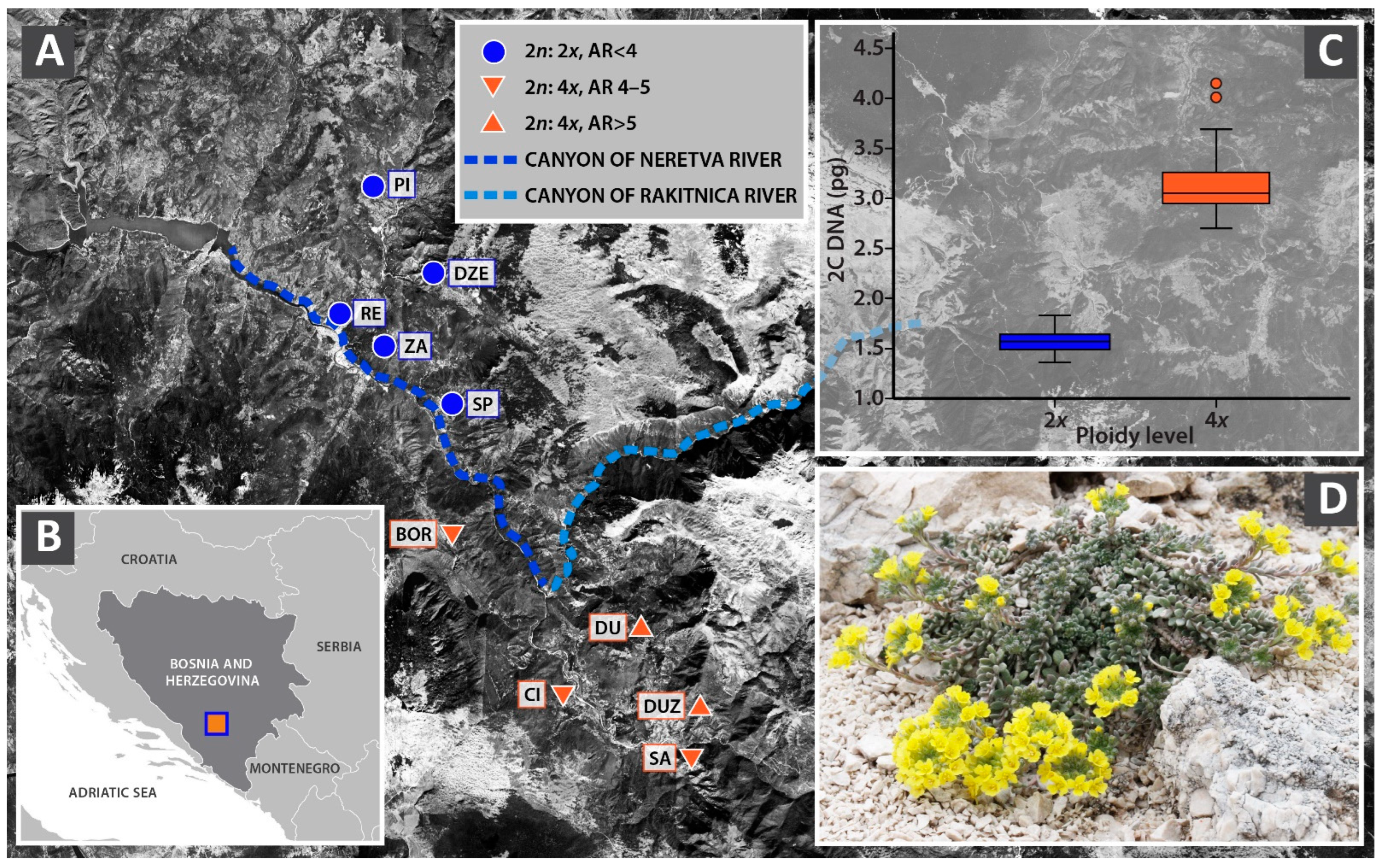

| 2C (pg) | 1Cx (pg) | 1Cx (Mbp) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Locality, ID | NFCM | Ploidy | Mean ± SD | Min–Max | CV (%) | Mean ± SD | Min–Max | CV (%) | Mean ± SD |
| Džepi, DZE | 19 | 2x | 1.60 ± 0.07 | 1.50–1.76 | 4.44 | 0.80 ± 0.04 | 0.75–0.88 | 4.60 | 784.38 ± 34.84 |
| Spiljani, SP | 20 | 2x | 1.64 ± 0.10 | 1.44–1.83 | 5.86 | 0.82 ± 0.05 | 0.72–0.91 | 5.67 | 803.79 ± 47.38 |
| Zagorice, ZA | 20 | 2x | 1.61 ± 0.07 | 1.36–1.70 | 4.64 | 0.81 ± 0.04 | 0.68–0.85 | 4.64 | 789.96 ± 36.77 |
| Pirići, PI | 20 | 2x | 1.51 ± 0.05 | 1.42–1.63 | 3.16 | 0.76 ± 0.02 | 0.71–0.82 | 3.28 | 740.11 ± 23.31 |
| Repovice, RE | 20 | 2x | 1.47 ± 0.04 | 1.38–1.60 | 2.98 | 0.74 ± 0.02 | 0.69–0.80 | 3.00 | 719.90 ± 21.26 |
| Čičevo, CI | 20 | 4x | 3.19 ± 0.17 | 2.70–3.44 | 5.38 | 0.80 ± 0.04 | 0.68–0.86 | 5.37 | 779.05 ± 41.79 |
| Sadine, SA | 20 | 4x | 3.23 ± 0.15 | 3.04–3.67 | 4.54 | 0.81 ± 0.04 | 0.76–0.92 | 4.62 | 790.11 ± 35.83 |
| Dudle, DU | 18 | 4x | 3.10 ± 0.20 | 2.80–3.51 | 6.52 | 0.77 ± 0.05 | 0.70–0.88 | 6.49 | 757.87 ± 49.33 |
| Borci, BOR | 20 | 4x | 2.96 ± 0.06 | 2.86–3.11 | 2.05 | 0.74 ± 0.02 | 0.72–0.78 | 2.09 | 724.02 ± 14.63 |
| Dužani, DUZ | 18 | 4x | 2.92 ± 0.10 | 2.79–3.14 | 3.52 | 0.73 ± 0.03 | 0.70–0.79 | 3.53 | 714.50 ± 25.27 |
| Ploidy | Locality ID | AN | AE | AR | R | p(R) | HE | HO | F |
|---|---|---|---|---|---|---|---|---|---|
| 2x | DZE | 3.0 | 1.71 | 3.00 | 0.586 | 0.506 | 0.369 | 0.260 | 0.301 |
| SP | 2.8 | 1.77 | 2.77 | 0.667 | 0.631 | 0.389 | 0.330 | 0.157 | |
| ZA | 3.4 | 2.47 | 3.40 | 0.725 | 0.563 | 0.494 | 0.350 | 0.192 | |
| PI | 4.0 | 2.54 | 4.00 | 0.620 | 0.566 | 0.462 | 0.300 | 0.276 | |
| RE | 3.6 | 2.52 | 3.60 | 0.694 | 0.557 | 0.509 | 0.350 | 0.200 | |
| 4x | CI | 4.6 | 2.84 | 4.45 | 0.612 | 0.224 | 0.601 | 0.640 | 0.129 |
| SA | 5.4 | 2.81 | 4.87 | 0.588 | 0.334 | 0.554 | 0.510 | 0.400 | |
| DU | 5.4 | 3.38 | 5.06 | 0.628 | 0.287 | 0.645 | 0.610 | 0.278 | |
| BOR | 4.4 | 2.75 | 4.18 | 0.649 | 0.299 | 0.564 | 0.500 | 0.314 | |
| DUZ | 6.0 | 3.51 | 5.48 | 0.578 | 0.120 | 0.665 | 0.550 | 0.343 |
| Loci | AN | AE | AR | R | p(R) | HE | HO | F | iMAF | p(iMAF) | FIT | FIS | FST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP34461 | 5 | 1.59 | 4.04 | 0.318 | 0.045 * | 0.369 | 0.338 | 0.342 | 0.255 | 0.007 * | 0.3467 | 0.2943 | 0.0744 |
| AP31679 | 7 | 3.49 | 5.46 | 0.499 | 0.017 * | 0.713 | 0.751 | −0.047 | 0.397 | 0.041 | −0.0397 | −0.1149 | 0.0675 |
| AP31733 | 6 | 2.26 | 3.91 | 0.377 | 0.044 * | 0.558 | 0.353 | 0.572 | 0.322 | 0.020 | 0.5777 | 0.5102 | 0.1377 |
| AP31640 | 10 | 1.68 | 6.45 | 0.168 | 0.003 * | 0.406 | 0.388 | 0.091 | 0.131 | 0.001 * | 0.1041 | −0.0450 | 0.1427 |
| AP10368 | 13 | 5.34 | 8.60 | 0.411 | 0.005 * | 0.813 | 0.318 | 0.718 | 0.265 | 0.009 * | 0.7211 | 0.6870 | 0.1089 |
| Average | 8.2 | 2.87 | 5.69 | 0.354 | 0.022 | 0.572 | 0.430 | 0.360 | 0.274 | 0.015 | 0.3668 | 0.2922 | 0.1055 |
| No. | Trait | PC1 | PC2 | DF1 |
|---|---|---|---|---|
| 1 | Stem length | 0.295 | −0.137 | 0.064 |
| 2 | 8th cauline leaf length | 0.345 | −0.204 | 0.006 |
| 3 | 8th cauline leaf width | 0.308 | −0.283 | - |
| 4 | 15th cauline leaf length | 0.332 | −0.306 | - |
| 5 | 15th cauline leaf width | 0.280 | −0.300 | - |
| 6 | 8th and 15th cauline leaf distance | 0.211 | 0.088 | - |
| 7 | Petal length | 0.285 | 0.346 | −0.210 |
| 8 | Petal width | 0.210 | 0.259 | - |
| 9 | Petal sinus deepness | 0.127 | 0.161 | −0.491 |
| 10 | Sepal length | 0.286 | 0.313 | −0.580 |
| 11 | Filament length | 0.318 | 0.272 | −0.428 |
| 12 | Style length | 0.252 | 0.311 | −0.443 |
| 13 | Trichome ray length on lower surface of middle cauline leaf | 0.177 | 0.060 | −0.038 |
| 14 | Trichome density on lower surface | −0.080 | 0.281 | −0.168 |
| 15 | Trichome coverage on lower surface | −0.049 | 0.006 | - |
| 16 | Trichome density on upper surface | −0.138 | 0.313 | 0.100 |
| 17 | Trichome coverage on upper surface | −0.129 | 0.075 | 0.239 |
| Diploids | Tetraploids | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI | ZA | SP | BOR | SA | DUŽ | |||||||
| P | E | P | E | P | E | P | E | P | E | P | E | |
| Min–Max | 27.5− 32.5 | 25.0− 27.5 | 27.5− 32.5 | 22.5− 30.0 | 25.0− 32.5 | 25.0− 27.5 | 32.5− 36.25 | 27.5− 32.5 | 35.0− 38.7 | 27.5− 31.5 | 35.0− 40.0 | 27.5− 32.5 |
| Mean ± SD | 30.9 ± 1.7 | 25.6 ± 1.1 | 29.9 ± 1.7 | 25.8 ± 1.9 | 29.6 ± 2.3 | 25.1 ± 1.3 | 34.7 ± 1.1 | 29.8 ± 1.0 | 36.7 ± 1.2 | 30.0 ± 0.7 | 37.5 ± 1.4 | 31.2 ± 1.5 |
| CV (%) | 5.6 | 4.1 | 5.7 | 7.2 | 7.7 | 5.1 | 3.1 | 3.5 | 3.4 | 2.3 | 3.7 | 4.8 |
| No. | Locality, ID | Latitude/Longitude | Altitude | NFCM | NSTR | NAFLP | Ncp | Nm | Voucher |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Čičevo, CI | 43.513780/18.080581 | 454 m | 20 | 20 | 5 | 4 | 20 | 53495 |
| 2. | Sadine, SA | 43.490831/18.148128 | 668 m | 20 | 20 | 5 | 5 | 20 | 53496 |
| 3. | Dudle, DU | 43.540567/18.121261 | 1034 m | 18 | 20 | 5 | 4 | 20 | 53497 |
| 4. | Borci, BOR | 43.573517/18.022017 | 711 m | 20 | 20 | 5 | 5 | 20 | 53503 |
| 5. | Dužani, DUZ | 43.509894/18.152114 | 830 m | 18 | 20 | 5 | 3 | 20 | 53504 |
| 6. | Džepi, DZE | 43.675506/18.011992 | 757 m | 19 | 20 | 5 | 4 | 19 | 53498 |
| 7. | Spiljani, SP | 43.625247/18.021678 | 509 m | 20 | 21 | 5 | 4 | 20 | 53499 |
| 8. | Zagorice, ZA | 43.647475/17.987317 | 668 m | 20 | 20 | 5 | 4 | 17 | 53500 |
| 9. | Pirići, PI | 43.708400/17.981864 | 623 m | 20 | 20 | 5 | 5 | 19 | 53501 |
| 10. | Repovice, RE | 43.659747/17.964069 | 338 m | 20 | 20 | 5 | 4 | 20 | 53502 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanjalić Kurtović, J.; Kalamujić Stroil, B.; Siljak-Yakovlev, S.; Pojskić, N.; Durmić-Pašić, A.; Hajrudinović-Bogunić, A.; Lasić, L.; Ušanović, L.; Bogunić, F. Spatial Distribution of Genetic, Ploidy, and Morphological Variation of the Edaphic Steno-Endemic Alyssum moellendorfianum (Brassicaceae) from the Western Balkans. Plants 2025, 14, 146. https://doi.org/10.3390/plants14020146
Hanjalić Kurtović J, Kalamujić Stroil B, Siljak-Yakovlev S, Pojskić N, Durmić-Pašić A, Hajrudinović-Bogunić A, Lasić L, Ušanović L, Bogunić F. Spatial Distribution of Genetic, Ploidy, and Morphological Variation of the Edaphic Steno-Endemic Alyssum moellendorfianum (Brassicaceae) from the Western Balkans. Plants. 2025; 14(2):146. https://doi.org/10.3390/plants14020146
Chicago/Turabian StyleHanjalić Kurtović, Jasna, Belma Kalamujić Stroil, Sonja Siljak-Yakovlev, Naris Pojskić, Adaleta Durmić-Pašić, Alma Hajrudinović-Bogunić, Lejla Lasić, Lejla Ušanović, and Faruk Bogunić. 2025. "Spatial Distribution of Genetic, Ploidy, and Morphological Variation of the Edaphic Steno-Endemic Alyssum moellendorfianum (Brassicaceae) from the Western Balkans" Plants 14, no. 2: 146. https://doi.org/10.3390/plants14020146
APA StyleHanjalić Kurtović, J., Kalamujić Stroil, B., Siljak-Yakovlev, S., Pojskić, N., Durmić-Pašić, A., Hajrudinović-Bogunić, A., Lasić, L., Ušanović, L., & Bogunić, F. (2025). Spatial Distribution of Genetic, Ploidy, and Morphological Variation of the Edaphic Steno-Endemic Alyssum moellendorfianum (Brassicaceae) from the Western Balkans. Plants, 14(2), 146. https://doi.org/10.3390/plants14020146








