On the Biosynthesis of Bioactive Tryptamines in Black Cohosh (Actaea racemosa L.)
Abstract
1. Introduction
2. Results
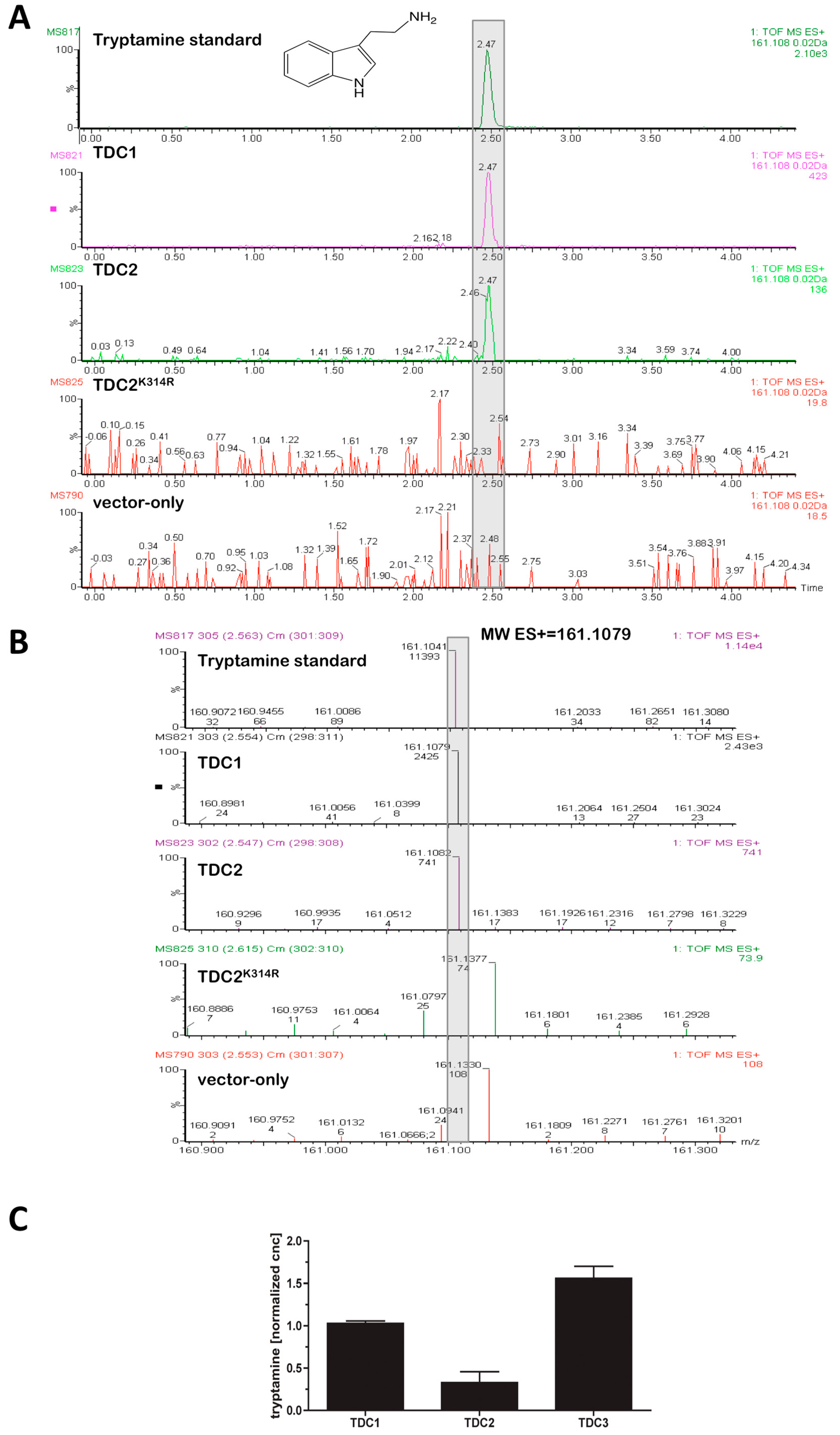
2.1. TDC1 and TDC2 Are PLP-Dependent Tryptophan Decarboxylases That Differ ~Four-Fold in Activity
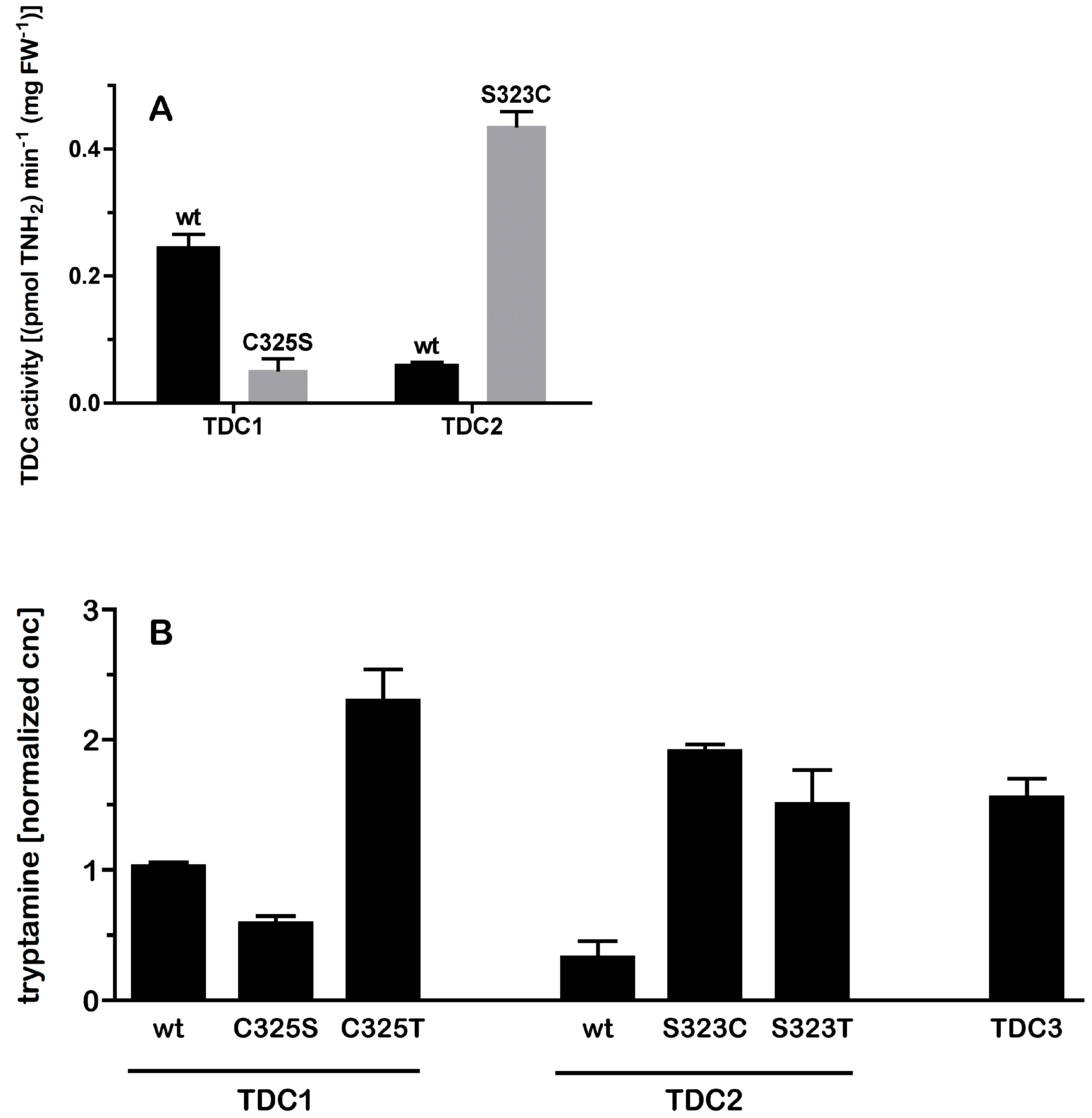
2.2. A Single Amino Acid Substitution Accounts for the Difference in Activity Between TDC2 and TDC1/TDC3
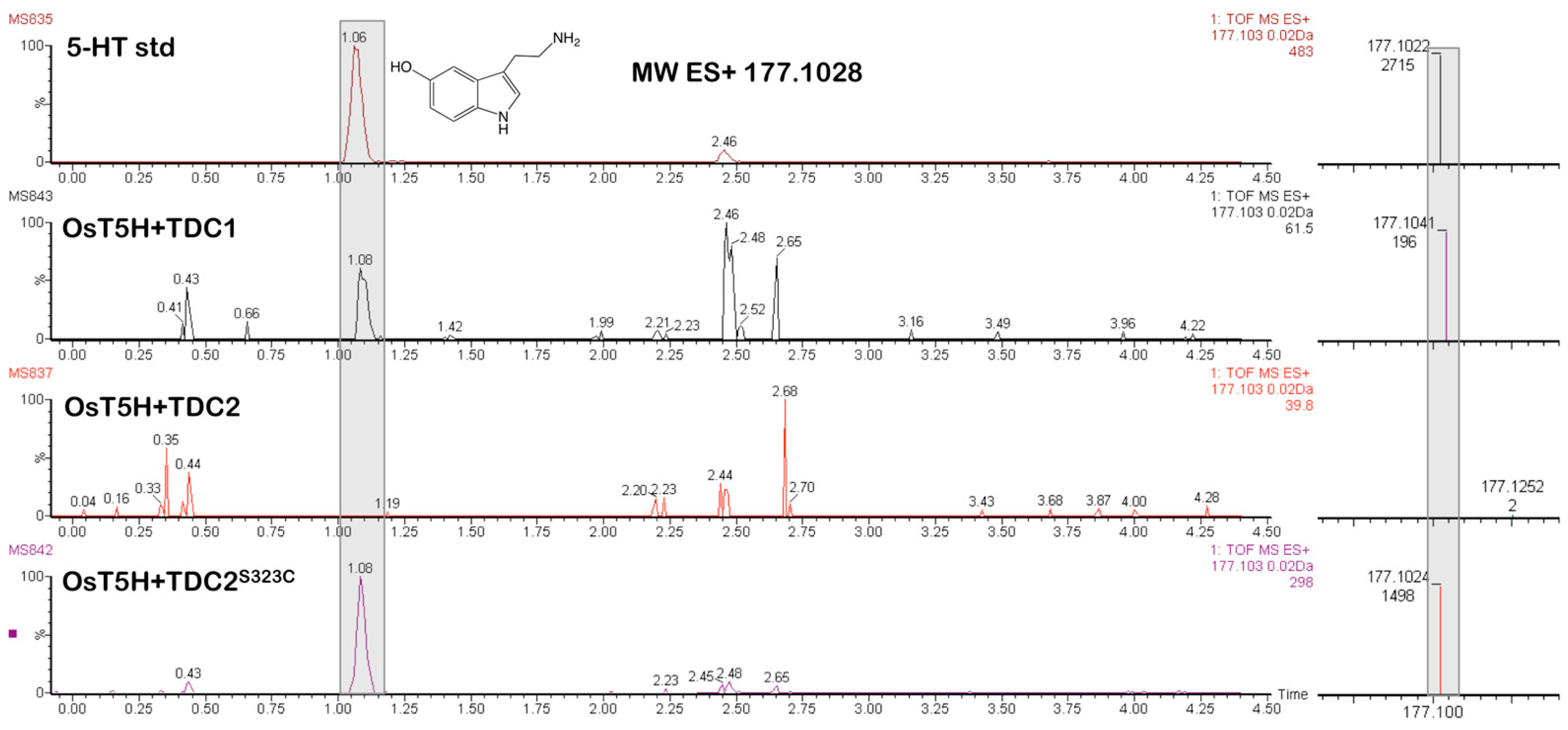
2.3. Co-Expression of TDCs from A. racemosa with Tryptamine-5-Hydroxylase (OsT5H) from Rice (O. sativa) Results in Serotonin Production
2.4. At Least Four Different Tryptamines Are Detectable in Black Cohosh
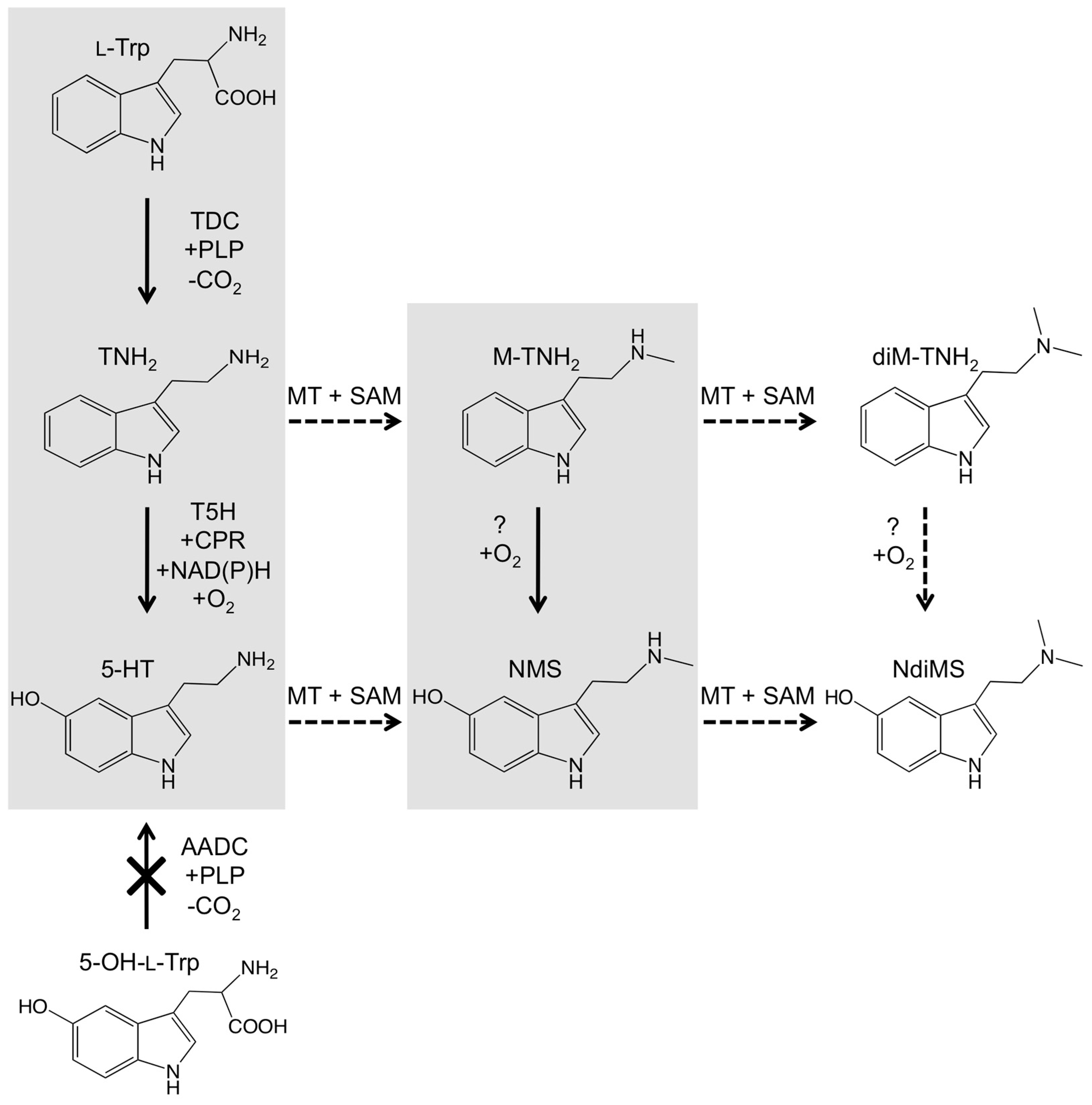
2.5. Feeding Experiments Suggest Biosynthetic Routes of Serotonin Metabolites in Black Cohosh
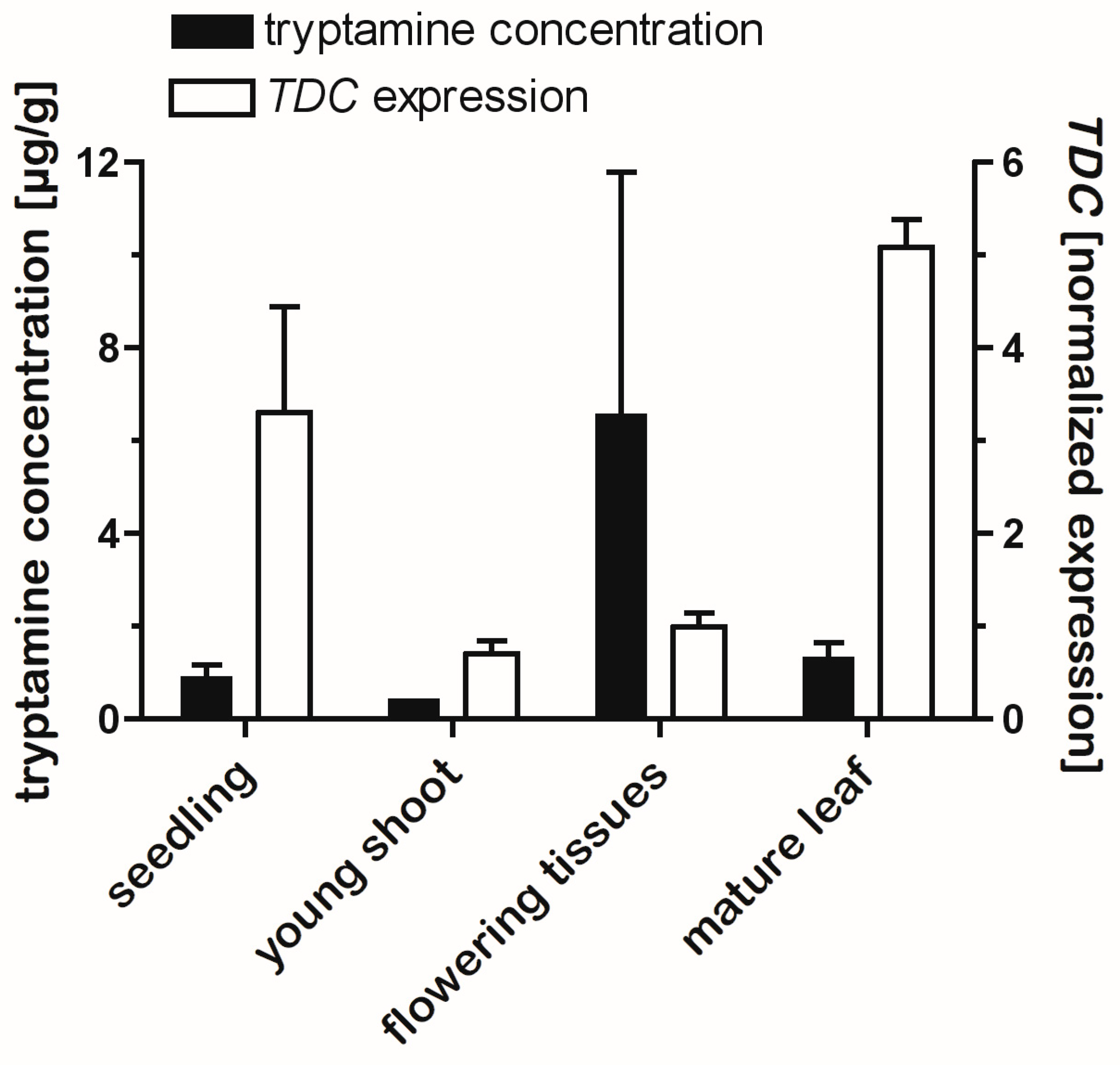
2.6. Tissue Levels of Tryptamines Are Only Weakly Correlated with TDC Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Chemicals, Strains, Enzymes, General Molecular Methods, and Biological Sequences
4.3. Functional Expression of Plant Genes in Microbial Hosts
4.4. Site-Directed Mutagenesis and Gene Fusions
4.5. Heterologous Expression in Yeast
4.6. Detached Leaf Assays for Precursor Feeding
4.7. Enzyme and Compound Extractions from Yeast Cells or Plant Tissues
4.8. UPLC-MS-TOF Analysis
4.9. Cloning of a Tryptamine-5-Hydroxylase Gene from Rice and of T5H-like and CPR Genes from Black Cohosh
4.10. Analysis of Gene Expression by Reverse-Transcription qPCR
4.11. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ma, C.; Kavalier, A.R.; Jiang, B.; Kennelly, E.J. Metabolic profiling of Actaea species extracts using high performance liquid chromatography coupled with electrospray ionization time-of-flight mass spectrometry. J. Chromatogr. A 2011, 1218, 1461–1476. [Google Scholar] [CrossRef]
- McCoy, J.A.; Davis, J.M.; Camper, N.D.; Khan, I.; Bharathi, A. Influence of rhizome propagule size on yields and triterpene glycoside concentrations of black cohosh [Actaea racemosa L. syn Cimicifuga racemosa (L.) Nuttal]. HortScience 2007, 42, 61–64. [Google Scholar] [CrossRef]
- Predny, M.L.; DeAngelis, P.; Chamberlain, J.L. Black Cohosh: An Annotated Bibliography; Southern Research Station: Asheville, NC, USA, 2006; pp. 1–108.
- Small, C.J.; Chamberlain, J.L.; Mathews, D.S. Recovery of black cohosh (Actaea racemosa L.) following experimental harvests. Am. Midl. Nat. 2011, 166, 339–348. [Google Scholar] [CrossRef]
- Castelo-Branco, C.; Gambacciani, M.; Cano, A.; Minkin, M.J.; Rachoń, D.; Ruan, X.; Beer, A.M.; Schnitker, J.; Henneicke-von Zepelin, H.H.; Pickartz, S. Review & meta-analysis: Isopropanolic black cohosh extract iCR for menopausal symptoms—An update on the evidence. Climacteric 2021, 24, 109–119. [Google Scholar] [PubMed]
- Castelo-Branco, C.; Navarro, C.; Beltrán, E.; Losa, F.; Camacho, M.; The Natural Products Study Group of the Spanish Menopause Society. Black cohosh efficacy and safety for menopausal symptoms. The Spanish Menopause Society statement. Gynecol. Endocrinol. 2022, 38, 379–384. [Google Scholar] [CrossRef]
- Fritz, H.; Seely, D.; McGowan, J.; Skidmore, B.; Fernandes, R.; Kennedy, D.A.; Cooley, K.; Wong, R.; Sagar, S.; Balneaves, L.G.; et al. Black cohosh and breast cancer: A systematic review. Integr. Cancer Ther. 2014, 13, 12–29. [Google Scholar] [CrossRef]
- Kligler, B. Black cohosh. Am. Fam. Physician 2003, 68, 114–116. [Google Scholar]
- Kretzschmar, G.; Nisslein, T.; Zierau, O.; Vollmer, G. No estrogen-like effects of an isopropanolic extract of Rhizoma Cimicifugae racemosae on uterus and vena cava of rats after 17 day treatment. J. Steroid Biochem. Mol. Biol. 2005, 97, 271–277. [Google Scholar] [CrossRef]
- Lupu, R.; Mehmi, I.; Atlas, E.; Tsai, M.S.; Pisha, E.; Oketch-Rabah, H.A.; Nuntanakorn, P.; Kennelly, E.J.; Kronenberg, F. Black cohosh, a menopausal remedy, does not have estrogenic activity and does not promote breast cancer cell growth. Int. J. Oncol. 2003, 23, 1407–1412. [Google Scholar] [CrossRef]
- Sadahiro, R.; Matsuoka, L.N.; Zeng, B.S.; Chen, K.H.; Zeng, B.Y.; Wang, H.Y.; Chu, C.S.; Stubbs, B.; Su, K.P.; Tu, Y.K.; et al. Black cohosh extracts in women with menopausal symptoms: An updated pairwise meta-analysis. Menopause 2023, 30, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Amato, P.; Marcus, D.M. Review of alternative therapies for treatment of menopausal symptoms. Climacteric 2003, 6, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, F.; Ernst, E. Black cohosh (Cimicifuga racemosa) for menopausal symptoms: A systematic review of its efficacy. Pharmacol. Res. 2008, 58, 8–14. [Google Scholar] [CrossRef]
- Cora, M. Black Cohosh Herbal Extract and Hematologic Alterations in B6C3F1/N Mice. Toxicol. Pathol. 2022, 50, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.L.; Fahey, J.W. Black cohosh: Coming full circle? J. Ethnopharmacol. 2012, 141, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Leach, M.J.; Moore, V. Black cohosh (Cimicifuga spp.) for menopausal symptoms. Cochrane Database Syst. Rev. 2012, 2012, CD007244. [Google Scholar] [CrossRef]
- Teschke, R.; Bahre, R.; Fuchs, J.; Wolff, A. Black cohosh hepatotoxicity: Quantitative causality evaluation in nine suspected cases. Menopause 2009, 16, 956–965. [Google Scholar] [CrossRef]
- Avula, B.; Ali, Z.; Khan, I.A. Chemical fingerprinting of Actaea racemosa (black cohosh) and its comparison study with closely related Actaea species (A. pachypoda, A. podocarpa, A. rubra) by HPLC. Chromatographia 2007, 66, 757–762. [Google Scholar] [CrossRef]
- Burdette, J.E.; Chen, S.N.; Lu, Z.Z.; Xu, H.; White, B.E.; Fabricant, D.S.; Liu, J.; Fong, H.H.; Farnsworth, N.R.; Constantinou, A.I.; et al. Black cohosh (Cimicifuga racemosa L.) protects against menadione-induced DNA damage through scavenging of reactive oxygen species: Bioassay-directed isolation and characterization of active principles. J. Agric. Food Chem. 2002, 50, 7022–7028. [Google Scholar] [CrossRef] [PubMed]
- Einbond, L.S.; Shimizu, M.; Ma, H.; Wu, H.A.; Goldsberry, S.; Sicular, S.; Panjikaran, M.; Genovese, G.; Cruz, E. Actein inhibits the Na+-K+-ATPase and enhances the growth inhibitory effect of digitoxin on human breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 375, 608–613. [Google Scholar] [CrossRef]
- Einbond, L.S.; Shimizu, M.; Xiao, D.; Nuntanakorn, P.; Lim, J.T.; Suzui, M.; Seter, C.; Pertel, T.; Kennelly, E.J.; Kronenberg, F.; et al. Growth inhibitory activity of extracts and purified components of black cohosh on human breast cancer cells. Breast Cancer Res. Treat. 2004, 83, 221–231. [Google Scholar]
- Jiang, B.; Ma, C.; Motley, T.; Kronenberg, F.; Kennelly, E.J. Phytochemical fingerprinting to thwart black cohosh adulteration: A 15 Actaea species analysis. Phytochem. Anal. 2011, 22, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Nuntanakorn, P.; Jiang, B.; Einbond, L.S.; Yang, H.; Kronenberg, F.; Weinstein, I.B.; Kennelly, E.J. Polyphenolic constituents of Actaea racemosa. J. Nat. Prod. 2006, 69, 314–318. [Google Scholar] [CrossRef]
- Qiu, S.X.; Dan, C.; Ding, L.S.; Peng, S.; Chen, S.N.; Farnsworth, N.R.; Nolta, J.; Gross, M.L.; Zhou, P. A triterpene glycoside from black cohosh that inhibits osteoclastogenesis by modulating RANKL and TNFalpha signaling pathways. Chem. Biol. 2007, 14, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Avula, B.; Wang, Y.H.; Smillie, T.J.; Khan, I.A. Quantitative determination of triterpenoids and formononetin in rhizomes of black cohosh (Actaea racemosa) and dietary supplements by using UPLC-UV/ELS detection and identification by UPLC-MS. Planta Med. 2009, 75, 381–386. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Pauli, G.F.; Zheng, B.; Wang, H.; Bai, N.; Peng, T.; Roller, M.; Zheng, Q. Cimicifuga species identification by high performance liquid chromatography-photodiode array/mass spectrometric/evaporative light scattering detection for quality control of black cohosh products. J. Chromatogr. A 2006, 1112, 241–254. [Google Scholar] [CrossRef]
- Li, W.; Chen, S.; Fabricant, D.; Angerhofer, C.K.; Fong, H.H.; Farnsworth, N.R.; Fitzloff, J.F. High-performance liquid chromatographic analysis of Black Cohosh (Cimicifuga racemosa) constituents with in-line evaporative light scattering and photodiode array detection. Anal. Chim. Acta 2002, 471, 61–75. [Google Scholar] [CrossRef]
- Shao, Y.; Harris, A.; Wang, M.; Zhang, H.; Cordell, G.A.; Bowman, M.; Lemmo, E. Triterpene glycosides from Cimicifuga racemosa. J. Nat. Prod. 2000, 63, 905–910. [Google Scholar] [CrossRef]
- Gödecke, T.; Nikolic, D.; Lankin, D.C.; Chen, S.N.; Powell, S.L.; Dietz, B.; Bolton, J.L.; Van Breemen, R.B.; Farnsworth, N.R.; Pauli, G.F. Phytochemistry of cimicifugic acids and associated bases in Cimicifuga racemosa root extracts. Phytochem. Anal. 2009, 20, 120–133. [Google Scholar] [CrossRef]
- Kruse, S.O.; Löhning, A.; Pauli, G.F.; Winterhoff, H.; Nahrstedt, A. Fukiic and piscidic acid esters from the rhizome of Cimicifuga racemosa and the in vitro estrogenic activity of fukinolic acid. Planta Med. 1999, 65, 763–764. [Google Scholar] [CrossRef]
- Li, W.; Sun, Y.; Liang, W.; Fitzloff, J.F.; van Breemen, R.B. Identification of caffeic acid derivatives in Actea racemosa (Cimicifuga racemosa, black cohosh) by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 978–982. [Google Scholar] [CrossRef]
- Cicek, S.S.; Khom, S.; Taferner, B.; Hering, S.; Stuppner, H. Bioactivity-guided isolation of GABA(A) receptor modulating constituents from the rhizomes of Actaea racemosa. J. Nat. Prod. 2010, 73, 2024–2028. [Google Scholar] [CrossRef]
- Nikolić, D.; Gödecke, T.; Chen, S.N.; White, J.; Lankin, D.C.; Pauli, G.F.; van Breemen, R.B. Mass spectrometric dereplication of nitrogen-containing constituents of black cohosh (Cimicifuga racemosa L.). Fitoterapia 2012, 83, 441–460. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.L.; Gödecke, T.; Nikolic, D.; Chen, S.N.; Ahn, S.; Dietz, B.; Farnsworth, N.R.; Van Breemen, R.B.; Lankin, D.C.; Pauli, G.F.; et al. In vitro serotonergic activity of black cohosh and identification of N(omega)-methylserotonin as a potential active constituent. J. Agric. Food Chem. 2008, 56, 11718–11726. [Google Scholar] [CrossRef]
- Burdette, J.E.; Liu, J.; Chen, S.N.; Fabricant, D.S.; Piersen, C.E.; Barker, E.L.; Pezzuto, J.M.; Mesecar, A.; Van Breemen, R.B.; Farnsworth, N.R.; et al. Black cohosh acts as a mixed competitive ligand and partial agonist of the serotonin receptor. J. Agric. Food Chem. 2003, 51, 5661–5670. [Google Scholar] [CrossRef]
- Spiering, M.J.; Urban, L.A.; Nuss, D.L.; Gopalan, V.; Stoltzfus, A.; Eisenstein, E. Gene identification in black cohosh (Actaea racemosa L.): Expressed sequence tag profiling and genetic screening yields candidate genes for production of bioactive secondary metabolites. Plant Cell Rep. 2011, 30, 613–629. [Google Scholar] [CrossRef]
- Facchini, P.J.; Huber-Allanach, K.L.; Tari, L.W. Plant aromatic L-amino acid decarboxylases: Evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 2000, 54, 121–138. [Google Scholar] [CrossRef]
- Fujiwara, T.; Maisonneuve, S.; Isshiki, M.; Mizutani, M.; Chen, L.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 2010, 285, 11308–11313. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell Rep. 2007, 26, 2009–2015. [Google Scholar] [CrossRef]
- Park, S.; Kang, K.; Lee, K.; Choi, D.; Kim, Y.S.; Back, K. Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta 2009, 230, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Herrick, D.; Peltz, S.W.; Jacobson, A. Measurements of mRNA decay rates. In Methods in Enzymology—Guide to Yeast Genetics and Molecular Biology; Guthrie, C., Fink, G.R., Eds.; Academic Press: New York, NY, USA, 1991; pp. 415–423. [Google Scholar]
- Park, S.; Kang, K.; Lee, S.W.; Ahn, M.J.; Bae, J.M.; Back, K. Production of serotonin by dual expression of tryptophan decarboxylase and tryptamine 5-hydroxylase in Escherichia coli. Appl. Microbiol. Biotechnol. 2011, 89, 1387–1394. [Google Scholar] [CrossRef]
- Urban, P.; Mignotte, C.; Kazmaier, M.; Delorme, F.; Pompon, D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 1997, 272, 19176–19186. [Google Scholar] [CrossRef]
- Ouwerkerk, P.B.; Memelink, J. Elicitor-responsive promoter regions in the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol. Biol. 1999, 39, 129–136. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef] [PubMed]
- Berlin, J.; Rugenhagen, C.; Dietze, P.; Fecker, L.F.; Goddijn, O.J.M.; Hoge, J.H.C. Increased production of serotonin by suspension and root cultures of Peganum harmala transformed with a tryptophan decarboxylase cDNA clone from Catharanthus roseus. Transgenic Res. 1993, 2, 336–344. [Google Scholar] [CrossRef]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 2007, 227, 263–272. [Google Scholar] [CrossRef]
- Glenn, W.S.; Nims, E.; O’Connor, S.E. Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J. Am. Chem. Soc. 2011, 133, 19346–19349. [Google Scholar] [CrossRef]
- Vigerelli, H.; Sciani, J.M.; Eula, M.A.C.; Sato, L.A.; Antoniazzi, M.M.; Jared, C.; Pimenta, D.C. Biological Effects and Biodistribution of Bufotenine on Mice. Biomed. Res. Int. 2018, 2018, 1032638. [Google Scholar] [CrossRef]
- Kang, K.; Park, S.; Natsagdorj, U.; Kim, Y.S.; Back, K. Methanol is an endogenous elicitor molecule for the synthesis of tryptophan and tryptophan-derived secondary metabolites upon senescence of detached rice leaves. Plant J. 2011, 66, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Schröder, P.; Abele, C.; Gohr, P.; Stuhlfauth-Roisch, U.; Grosse, W. Latest on enzymology of serotonin biosynthesis in walnut seeds. Adv. Exp. Med. Biol. 1999, 467, 637–644. [Google Scholar]
- Compton, J.; Culham, A.; Jury, S. Reclassification of Actaea to include Cimicifuga and Souliea (Ranunculaceae): Phylogeny inferred from morphology, nrDNA ITS, and cpDNA trnL-F sequence variation. Taxon 1998, 47, 593–634. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Register, E.; Curotto, J.; Kurtz, M.; Kelly, R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 1998, 14, 565–571. [Google Scholar] [CrossRef]
- Gollub, E.G.; Liu, K.P.; Dayan, J.; Adlersberg, M.; Sprinson, D.B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 1977, 252, 2846–2854. [Google Scholar] [CrossRef]
- Lopez-Meyer, M.; Nessler, C.L. Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J. 1997, 11, 1167–1175. [Google Scholar] [CrossRef] [PubMed]



| Concentration in µg gFW−1 | ||||
|---|---|---|---|---|
| Mean (Range) a | ||||
| Plant tissue b | TNH2 | 5-HT | NMS | NdiMS |
| Rhizome | 0.0 b | 3.7 (0.0–5.9) | 5.5 (0.3–9.6) | trace (NA) |
| Root | 0.0 | 0.9 (0.0–1.5) | 4.1 (0.3–9.6) | 0.0 |
| Young leaf | 0.0 | 0.8 (0.0–1.7) | 0.8 (0.7–1.1) | 0.0 |
| Mature leaf | 0.0 | 1.8 (0.0–6.9) | 1.3 (0.5–4.2) | 0.7 (0.0–0.7) |
| Flower buds | 0.2 (0.0–0.4) | 4.9 (0.0–8.6) | 1.4 (0.0–2.7) | 1.7 (0.0–3.8) |
| Mature flowers | 0.9 (0.0–0.9) | 8.6 (1.2–21.6) | 4.2 (0.5–11.2) | 4.4 (0.0–8.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spiering, M.J.; Parsons, J.F.; Eisenstein, E. On the Biosynthesis of Bioactive Tryptamines in Black Cohosh (Actaea racemosa L.). Plants 2025, 14, 292. https://doi.org/10.3390/plants14020292
Spiering MJ, Parsons JF, Eisenstein E. On the Biosynthesis of Bioactive Tryptamines in Black Cohosh (Actaea racemosa L.). Plants. 2025; 14(2):292. https://doi.org/10.3390/plants14020292
Chicago/Turabian StyleSpiering, Martin J., James F. Parsons, and Edward Eisenstein. 2025. "On the Biosynthesis of Bioactive Tryptamines in Black Cohosh (Actaea racemosa L.)" Plants 14, no. 2: 292. https://doi.org/10.3390/plants14020292
APA StyleSpiering, M. J., Parsons, J. F., & Eisenstein, E. (2025). On the Biosynthesis of Bioactive Tryptamines in Black Cohosh (Actaea racemosa L.). Plants, 14(2), 292. https://doi.org/10.3390/plants14020292








