Multiprotein Complexes of Plant Glycosyltransferases Involved in Their Function and Trafficking
Abstract
:1. Introduction
2. Protein-Protein Interactions Among GTs Involved in Polysaccharide Synthesis and Protein Glycosylation in Golgi
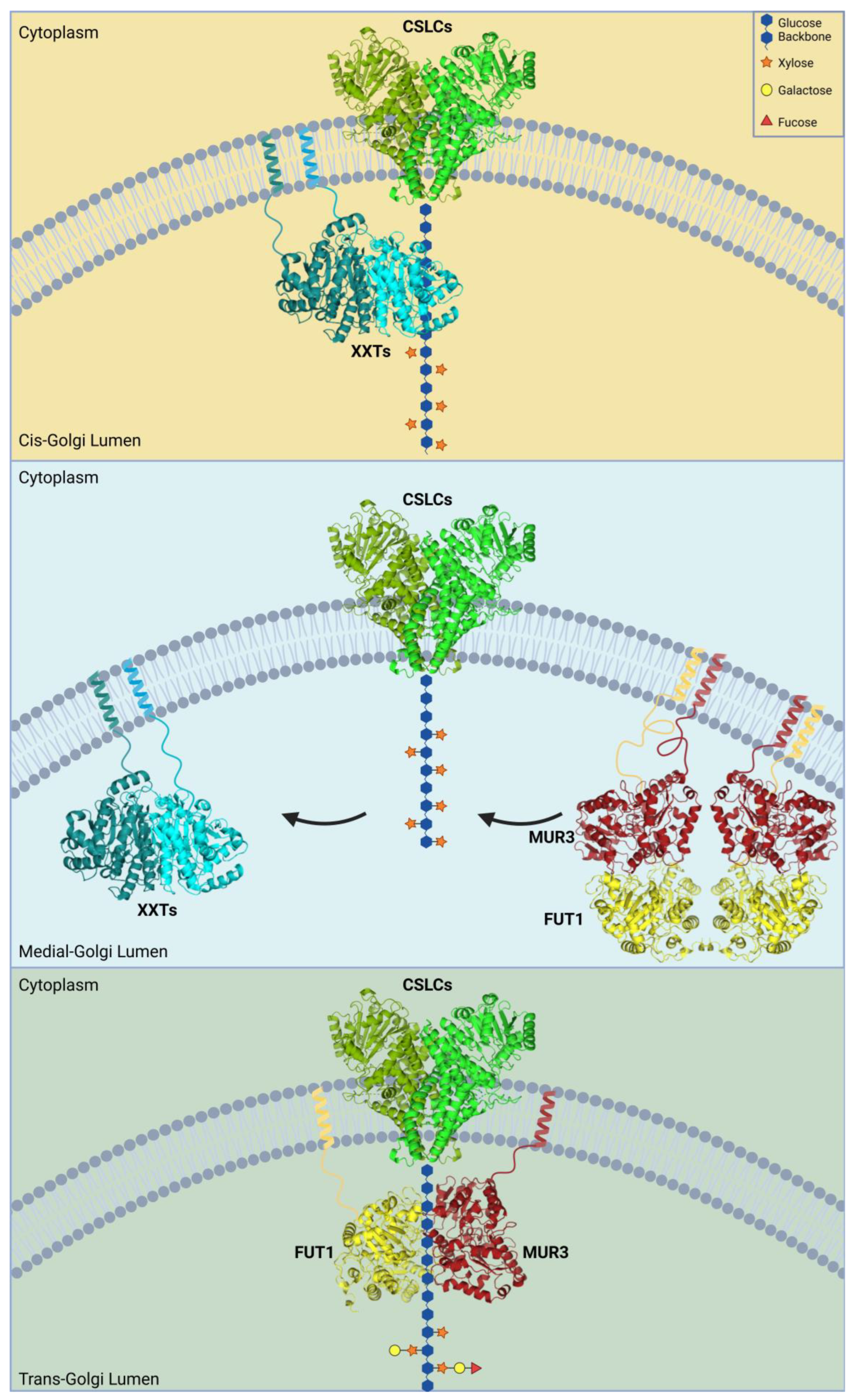
2.1. Xyloglucan Synthase Complex
| Xyloglucan Synthase Complex | |||
| Protein | Heterodimer With | Homodimer | |
| AtCSLC4 | AtXXT2, AtXXT5, AtXLT2, AtMUR3, AtFUT1 |  | [50,59,60] |
| AtXXT1 | AtXXT2, AtXXT5 |  | [50,59,60,63] |
| AtXXT2 | AtCSLC4, AtXXT1, AtXXT5, AtXLT2, AtFUT1 |  | [50,59,60] |
| AtXXT5 | AtCSLC4, AtXXT1, AtXXT2, AtXLT2, AtFUT1 | [50,59,60] | |
| AtXLT2 | AtCSLC4, AtXXT2, AtXXT5, AtFUT1 | [59,60] | |
| AtMUR3 | AtCSLC4, AtFUT1 | [59,60] | |
| AtFUT1 | AtCSLC4, AtXXT2, AtXXT5, AtXLT2, AtMUR3 |  | [59,60,64,65] |
| Xylan Synthase Complex | |||
| Protein | Heterodimer With | Homodimer | |
| AoIRX9 | AoIRX14 |  | [66] |
| AoIRX10 |  | [66] | |
| AoIRX14 | AoIRX9 |  | [66] |
| TaGT43-4 | TaGT47-13, TaGLP, TaVER2, TaGT75-3, TaGT75-4 |  | [67] |
| TaGLP | TaGT43-4 |  | [67] |
| TaGT75-3 | TaGT43-4 |  | [67] |
| TaGT75-4 | TaGT43-4 |  | [67] |
| OsGT43B | OsGT47-2 | [68] | |
| OsGT43F | OsGT47-4 |  | [68] |
| OsGT43I | OsGT47-1, OsGT47-3 |  | [68] |
| OsGT43J | OsGT47-2 |  | [68] |
| Pectin Synthase Complex | |||
| Protein | Heterodimer With | Homodimer | |
| AtGAUT1 | AtGAUT5, AtGAUT6, AtGAUT7 |  | [69,70,71] |
| AtGAUT10, ATGAUT13, AtGAUT14 |  | [70] | |
| Arabinogalactan synthesizing GTs | |||
| Protein | Heterodimer With | Homodimer | |
| AtARAD1 | AtARAD1 |  | [72] |
| AtARAD2 | AtARAD2 |  | [72] |
| NaARAD1 | AtARAD1 |  | [73] |
| AtGALT29A | AtGALT14A, AtGALT31A |  | [74] |
| AtGALT31A |  | [74] | |
| Proteins involved in N-glycosylation | |||
| Protein | Heterodimer With | Homodimer | |
| GNTI | MNSI |  | [75,76] |
| XYLT | GMII |  | [75,76] |
| GALTI | GMII | [75] | |
 check mark indicates the confirmation of protein homodimer.
check mark indicates the confirmation of protein homodimer.2.2. Xylan Synthase Complex
2.3. Pectin Synthase Complex
2.4. Arabinogalactan Synthesizing GTs
2.5. Proteins Involved in N-glycosylation
2.6. Structural Analysis of the Homo- and Heterodimerization of GTs
3. The Trafficking of GTs Between ER and Golgi and Their Interaction with Coat Proteins
4. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cantarel, B.I.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyl Transferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Zabotina, O.A.; Zhang, N.; Weerts, R. Polysaccharide Biosynthesis: Glycosyltransferases and Their Complexes. Front. Plant Sci. 2021, 12, 625307. [Google Scholar] [CrossRef]
- Kumar, M.; Turner, S. Plant Cellulose Synthesis: CESA Proteins Crossing Kingdoms. Phytochemistry 2015, 112, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Structure and Growth of Plant Cell Walls. Nat. Rev. Mol. Cell Biol. 2024, 25, 340–358. [Google Scholar] [CrossRef]
- Delmer, D.; Dixon, R.A.; Keegstra, K.; Mohnen, D. The Plant Cell Wall-Dynamic, Strong, and Adaptable-Is a Natural Shapeshifter. Plant Cell 2024, 36, 1257–1311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The Plant Cell Wall: Biosynthesis, Construction, and Functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Cui, D.; Ye, Z.H. Secondary Cell Wall Biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.T. We Be Jammin’: An Update on Pectin Biosynthesis, Trafficking and Dynamics. J. Exp. Bot. 2016, 67, 495–502. [Google Scholar] [CrossRef]
- Julian, J.D.; Zabotina, O.A. Xyloglucan Biosynthesis: From Genes to Proteins and Their Functions. Front. Plant Sci. 2022, 13, 920494. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin Structure and Biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Zabotina, O.A. Xyloglucan and Its Biosynthesis. Front. Plant Sci. 2012, 3, 134. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- van Tol, W.; Wessels, H.; Lefeber, D.J. O-Glycosylation Disorders Pave the Road for Understanding the Complex Human O-Glycosylation Machinery. Curr. Opin. Struct. Biol. 2019, 56, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Van Den Steen, P.; Rudd, P.M.; Dwek, R.A.; Opdenakker, G. Concepts and Principles of O-Linked Glycosylation. Crit. Rev. Biochem. Mol. Biol. 1998, 33, 151–208. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.J.; Narimatsu, Y.; Schjoldager, K.T.; Tytgat, H.L.P.; Aebi, M.; Clausen, H.; Halim, A. SnapShot: O-Glycosylation Pathways across Kingdoms. Cell 2018, 172, 632–632.e2. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.L.; MacAlister, C.A.; Ulvskov, P. Plant Protein O-Arabinosylation. Front. Plant Sci. 2021, 12, 645219. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Seifert, G.; Doblin, M.S.; Johnson, K.L.; Ruprecht, C.; Pfrengle, F.; Bacic, A.; Estevez, J.M. Cracking the “Sugar Code”: A Snapshot of N- and O-Glycosylation Pathways and Functions in Plants Cells. Front. Plant Sci. 2021, 12, 640919. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.H.; Yang, B.; D’Souza, A.K.; Shen, D.; Woo, C.M. Truncation of the TPR Domain of OGT Alters Substrate and Glycosite Selection. Anal. Bioanal. Chem. 2021, 413, 7385–7399. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wei, X.; Cong, J.; Zou, J.; Wan, L.; Xu, S. Structural Insights into Mechanism and Specificity of the Plant Protein O-Fucosyltransferase SPINDLY. Nat. Commun. 2022, 13, 7424. [Google Scholar] [CrossRef]
- Aizezi, Y.; Zhao, H.; Zhang, Z.; Bi, Y.; Yang, Q.; Guo, G.; Zhang, H.; Guo, H.; Jiang, K.; Wang, Z.Y. Structure-Based Virtual Screening Identifies Small-Molecule Inhibitors of O-Fucosyltransferase SPINDLY in Arabidopsis. Plant Cell 2024, 36, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shi, X.; Yang, C.; Zhao, X.; Zhuang, J.; Liu, Y.; Gao, L.; Xia, T. Two UDP-Glycosyltransferases Catalyze the Biosynthesis of Bitter Flavonoid 7-O-Neohesperidoside through Sequential Glycosylation in Tea Plants. J. Agric. Food Chem. 2022, 70, 2354–2365. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Feng, X.; Ono, N.N.; Holland, D.; Amir, R.; Tian, L. Characterization of a UGT84 Family Glycosyltransferase Provides New Insights into Substrate Binding and Reactivity of Galloylglucose Ester-Forming UGTs. Biochemistry 2017, 56, 6389–6400. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, G.; Chang, Z.; Tang, Y.; Gao, H.; Pang, Y. Involvement of Three Putative Glucosyltransferases from the UGT72 Family in Flavonol Glucoside/Rhamnoside Biosynthesis in Lotus Japonicus Seeds. J. Exp. Bot. 2017, 68, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, Y.; von Schaewen, A.; Koiwa, H. Function of N-Glycosylation in Plants. Plant Sci. 2018, 274, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Plant Protein Glycosylation. Glycobiology 2016, 26, 926–939. [Google Scholar] [CrossRef]
- Aebi, M. N-Linked Protein Glycosylation in the ER. Biochim. Biophys. Acta. Mol. Cell Res. 2013, 1833, 2430–2437. [Google Scholar] [CrossRef]
- Liebminger, E.; Hüttner, S.; Vavra, U.; Fischl, R.; Schoberer, J.; Grass, J.; Blaukopf, C.; Seifert, G.J.; Altmann, F.; Mach, L.; et al. Class I α-Mannosidases Are Required for N-Glycan Processing and Root Development in Arabidopsis Thaliana. Plant Cell 2009, 21, 3850–3867. [Google Scholar] [CrossRef] [PubMed]
- Boisson, M.; Gomord, V.; Audran, C.; Berger, N.; Dubreucq, B.; Granier, F.; Lerouge, P.; Faye, L.; Caboche, M.; Lepiniec, L. Arabidopsis Glucosidase I Mutants Reveal a Critical Role of N-Glycan Trimming in Seed Development. EMBO J. 2001, 20, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Liebminger, E.; Christiane, V.; Mach, L.; Strasser, R. Mannose Trimming Reactions in the Early Stages of the N-Glycan Pathway. Plant Signal Behav. 2010, 5, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Von Schaewen, A.; Sturm, A.; O’neill, J.; Chrispeels, M.J. Isolation of a Mutant Arabidopsis Plant that Lacks N-acetyl Glucosaminyl Transferase I and Is Unable to Synthesize Golgi-modified Complex N-linked Glycans. Plant Physiol. 1993, 102, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Altmann, F.; Mach, L.; Glössl, J.; Steinkellner, H. Generation of Arabidopsis Thaliana Plants with Complex N-Glycans Lacking Β1,2-Linked Xylose and Core A1,3-Linked Fucose. FEBS Lett. 2004, 561, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Kajiura, H.; Okamoto, T.; Misaki, R.; Matsuura, Y.; Fujiyama, K. Arabidopsis Β1,2-Xylosyltransferase: Substrate Specificity and Participation in the Plant-Specific N-Glycosylation Pathway. J. Biosci. Bioeng. 2012, 113, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Leiter, H.; Mucha, J.; Staudacher, E.; Grimm, R.; Glössl, J.; Altmann, F. Purification, CDNA Cloning, and Expression of GDP-L-Fuc:Asn-Linked GlcNAc A1,3-Fucosyltransferase from Mung Beans. J. Biol. Chem. 1999, 274, 21830–21839. [Google Scholar] [CrossRef]
- Strasser, R.; Mucha, J.; MacH, L.; Altmann, F.; Wilson, I.B.H.; Glössl, J.; Steinkellner, H. Molecular Cloning and Functional Expression of Β1,2-Xylosyltransferase CDNA from Arabidopsis Thaliana. FEBS Lett. 2000, 472, 105–108. [Google Scholar] [CrossRef]
- Wilson, I.B.H.; Zeleny, R.; Kolarich, D.; Staudacher, E.; Stroop, C.J.M.; Kamerling, J.P.; Altmann, F. Analysis of Asn-linked Glycans from Vegetable Foodstuffs: Widespread Occurrence of Lewis A, Core Alpha1,3-linked Fucose and Xylose Substitutions. Glycobiology 2001, 11, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Bondili, J.S.; Vavra, U.; Schoberer, J.; Svoboda, B.; Glössl, J.; Léonard, R.; Stadlmann, J.; Altmann, F.; Steinkellner, H.; et al. A Unique Β1,3-Galactosyltransferase Is Indispensable for the Biosynthesis of N-Glycans Containing Lewis a Structures in Arabidopsis Thaliana. Plant Cell 2007, 19, 2278–2292. [Google Scholar] [CrossRef]
- Allen, H.; Wei, D.; Gu, Y.; Li, S. A Historical Perspective on the Regulation of Cellulose Biosynthesis. Carbohydr. Polym. 2021, 252, 117022. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, G.B.; Blaschek, L.; Frandsen, K.E.H.; Noack, L.C.; Persson, S. Cellulose Synthesis in Land Plants. Mol. Plant 2023, 16, 206–231. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Keegstra, K. Biosynthesis of the Plant Cell Wall Matrix Polysaccharide Xyloglucan. Annu. Rev. Plant Biol. 2016, 67, 235–259. [Google Scholar] [CrossRef]
- Hsieh, Y.S.Y.; Harris, P.J. Xyloglucans of Monocotyledons Have Diverse Structures. Mol. Plant 2009, 2, 943–965. [Google Scholar] [CrossRef] [PubMed]
- Peña, M.J.; Darvill, A.G.; Eberhard, S.; York, W.S.; O’Neill, M.A. Moss and Liverwort Xyloglucans Contain Galacturonic Acid and Are Structurally Distinct from the Xyloglucans Synthesized by Hornworts and Vascular Plants. Glycobiology 2008, 18, 891–904. [Google Scholar] [CrossRef]
- Hsieh, Y.S.Y.; Harris, P.J. Structures of Xyloglucans in Primary Cell Walls of Gymnosperms, Monilophytes (Ferns Sensu Lato) and Lycophytes. Phytochemistry 2012, 79, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.; Jia, Z.; Peña, M.J.; Cash, M.; Harper, A.; Blackburn, A.R.; Darvill, A.; York, W.S. Structural Analysis of Xyloglucans in the Primary Cell Walls of Plants in the Subclass Asteridae. Carbohydr. Res. 2005, 340, 1826–1840. [Google Scholar] [CrossRef] [PubMed]
- Schultink, A.; Liu, L.; Zhu, L.; Pauly, M. Structural Diversity and Function of Xyloglucan Sidechain Substituents. Plants 2014, 3, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C.; York, W.S.; Albersheim, P.; Darvill, A.; Hayashi, T.; Joseleau, J.-P.; Kato, Y.; Lorences, E.P.; Maclachlan, G.A.; McNeil, M.; et al. An Unambiguous Nomenclature for Xyloglucan-derived Oligosaccharides. Physiol. Plant 1993, 89, 1–3. [Google Scholar] [CrossRef]
- Tuomivaara, S.T.; Yaoi, K.; O’Neill, M.A.; York, W.S. Generation and Structural Validation of a Library of Diverse Xyloglucan-Derived Oligosaccharides, Including an Update on Xyloglucan Nomenclature. Carbohydr. Res. 2015, 402, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Peña, M.J.; Kong, Y.; York, W.S.; O’Neill, M.A. A Galacturonic Acid-Containing Xyloglucan Is Involved in Arabidopsis Root Hair Tip Growth. Plant Cell 2012, 24, 4511–4524. [Google Scholar] [CrossRef] [PubMed]
- Cocuron, J.-C.; Lerouxel, O.; Drakakaki, G.; Alonso, A.P.; Liepman, A.H.; Keegstra, K.; Raikhel, N.; Wilkerson, C.G. A Gene from the Cellulose Synthase-like C Family Encodes a Beta-1,4 Glucan Synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 8550–8555. [Google Scholar] [CrossRef]
- Chou, Y.H.; Pogorelko, G.; Zabotina, O.A. Xyloglucan Xylosyltransferases XXT1, XXT2, and XXT5 and the Glucan Synthase CSLC4 Form Golgi-Localized Multiprotein Complexes. Plant Physiol. 2012, 159, 1355–1366. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Chandrasekar, B.; Rea, A.C.; Danhof, L.; Zemelis-Durfee, S.; Thrower, N.; Shepard, Z.S.; Pauly, M.; Brandizzi, F.; Keegstra, K. The Synthesis of Xyloglucan, an Abundant Plant Cell Wall Polysaccharide, Requires CSLC Function. Proc. Natl. Acad. Sci. USA 2020, 117, 20316–20324. [Google Scholar] [CrossRef] [PubMed]
- Faik, A.; Price, N.J.; Raikhel, N.V.; Keegstra, K. An Arabidopsis Gene Encoding An-Xylosyltransferase Involved in Xyloglucan Biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 7797–7802. [Google Scholar] [CrossRef] [PubMed]
- Cavalier, D.M.; Keegstra, K. Two Xyloglucan Xylosyltransferases Catalyze the Addition of Multiple Xylosyl Residues to Cellohexaose. J. Biol. Chem. 2006, 281, 34197–34207. [Google Scholar] [CrossRef] [PubMed]
- Zabotina, O.A.; Avci, U.; Cavalier, D.; Pattathil, S.; Chou, Y.H.; Eberhard, S.; Danhof, L.; Keegstra, K.; Hahn, M.G. Mutations in Multiple XXT Genes of Arabidopsis Reveal the Complexity of Xyloglucan Biosynthesis. Plant Physiol. 2012, 159, 1367–1384. [Google Scholar] [CrossRef]
- Culbertson, A.T.; Chou, Y.H.; Smith, A.L.; Young, Z.T.; Tietze, A.A.; Cottaz, S.; Fauré, R.; Zabotina, O.A. Enzymatic Activity of Xyloglucan Xylosyltransferase 5. Plant Physiol. 2016, 171, 1893–18904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Julian, J.D.; Yap, C.E.; Swaminathan, S.; Zabotina, O.A. The Arabidopsis Xylosyltransferases, XXT3, XXT4, and XXT5 Are Essential to Complete the Fully Xylosylated Glucan Backbone XXXG-type Structure of Xyloglucans. New Phytol. 2023, 238, 1986–1999. [Google Scholar] [CrossRef]
- Madson, M.; Dunand, C.; Li, X.; Verma, R.; Vanzin, G.F.; Caplan, J.; Shoue, D.A.; Carpita, N.C.; Reiter, W.D. The MUR3 Gene of Arabidopsis Encodes a Xyloglucan Galactosyltransferase That Is Evolutionarily Related to Animal Exostosins. Plant Cell 2003, 15, 1662–1670. [Google Scholar] [CrossRef]
- Jensen, J.K.; Schultink, A.; Keegstra, K.; Wilkerson, C.G.; Pauly, M. RNA-Seq Analysis of Developing Nasturtium Seeds (Tropaeolum Majus): Identification and Characterization of an Additional Galactosyltransferase Involved in Xyloglucan Biosynthesis. Mol. Plant 2012, 5, 984–992. [Google Scholar] [CrossRef]
- Chou, Y.H.; Pogorelko, G.; Young, Z.T.; Zabotina, O.A. Protein-Protein Interactions among Xyloglucan-Synthesizing Enzymes and Formation of Golgi-Localized Multiprotein Complexes. Plant Cell Physiol. 2015, 56, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Lund, C.H.; Bromley, J.R.; Stenbæk, A.; Rasmussen, R.E.; Scheller, H.V.; Sakuragi, Y. A Reversible Renilla Luciferase Protein Complementation Assay for Rapid Identification of Protein-Protein Interactions Reveals the Existence of an Interaction Network Involved in Xyloglucan Biosynthesis in the Plant Golgi Apparatus. J. Exp. Bot. 2015, 66, 85–97. [Google Scholar] [CrossRef]
- Chevalier, L.; Bernard, S.; Ramdani, Y.; Lamour, R.; Bardor, M.; Lerouge, P.; Follet-Gueye, M.L.; Driouich, A. Subcompartment Localization of the Side Chain Xyloglucan-Synthesizing Enzymes within Golgi Stacks of Tobacco Suspension-Cultured Cells. Plant J. 2010, 64, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.T.; Stevens, T.J.; McFarlane, H.E.; Vidal-Melgosa, S.; Griss, J.; Lawrence, N.; Butler, R.; Sousa, M.M.L.; Salemi, M.; Willats, W.G.T.; et al. Separating Golgi Proteins from Cis to Trans Reveals Underlying Properties of Cisternal Localization. Plant Cell 2019, 31, 2010–2034. [Google Scholar] [CrossRef] [PubMed]
- Culbertson, A.T.; Ehrlich, J.J.; Choe, J.Y.; Honzatko, R.B.; Zabotina, O.A. Structure of Xyloglucan Xylosyltransferase 1 Reveals Simple Steric Rules That Define Biological Patterns of Xyloglucan Polymers. Proc. Natl. Acad. Sci. USA 2018, 115, 6064–6069. [Google Scholar] [CrossRef] [PubMed]
- Urbanowicz, B.R.; Bharadwaj, V.S.; Alahuhta, M.; Peña, M.J.; Lunin, V.V.; Bomble, Y.J.; Wang, S.; Yang, J.Y.; Tuomivaara, S.T.; Himmel, M.E.; et al. Structural, Mutagenic and in Silico Studies of Xyloglucan Fucosylation in Arabidopsis Thaliana Suggest a Water-Mediated Mechanism. Plant J. 2017, 91, 931–949. [Google Scholar] [CrossRef]
- Rocha, J.; Cicéron, F.; de Sanctis, D.; Lelimousin, M.; Chazalet, V.; Lerouxel, O.; Breton, C. Structure of Arabidopsis Thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation. Plant Cell 2016, 28, 2352–2364. [Google Scholar] [CrossRef]
- Zeng, W.; Lampugnani, E.R.; Picard, K.L.; Song, L.; Wu, A.M.; Farion, I.M.; Zhao, J.; Ford, K.; Doblin, M.S.; Bacic, A. Asparagus IRX9, IRX10, and IRX14A Are Components of an Active Xylan Backbone Synthase Complex That Forms in the Golgi Apparatus. Plant Physiol. 2016, 171, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wiemels, R.E.; Soya, A.; Whitley, R.; Held, M.; Faik, A. Composition, Assembly, and Trafficking of a Wheat Xylan Synthase Complex. Plant Physiol. 2016, 170, 1999–2023. [Google Scholar] [CrossRef]
- Javaid, T.; Bhattarai, M.; Venkataraghavan, A.; Held, M.; Faik, A. Specific Protein Interactions between Rice Members of the GT43 and GT47 Families Form Various Central Cores of Putative Xylan Synthase Complexes. Plant J. 2024, 118, 856–878. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Sakuragi, Y.; Zhu, X.; Burrell, A.J.; Mohanty, S.S.; Atwood, J.A.; Orlando, R.; Scheller, H.V.; Mohnen, D. Galacturonosyltransferase (GAUT)1 and GAUT7 Are the Core of a Plant Cell Wall Pectin Biosynthetic Homogalacturonan:Galacturonosyltransferase Complex. Proc. Natl. Acad. Sci. USA 2011, 108, 20225–20230. [Google Scholar] [CrossRef]
- Engle, K.A.; Amos, R.A.; Yang, J.Y.; Glushka, J.; Atmodjo, M.A.; Tan, L.; Huang, C.; Moremen, K.W.; Mohnen, D. Multiple Arabidopsis Galacturonosyltransferases Synthesize Polymeric Homogalacturonan by Oligosaccharide Acceptor-Dependent or de Novo Synthesis. Plant J. 2022, 109, 1441–1456. [Google Scholar] [CrossRef] [PubMed]
- Lund, C.H.; Stenbæk, A.; Atmodjo, M.A.; Rasmussen, R.E.; Moller, I.E.; Erstad, S.M.; Biswal, A.K.; Mohnen, D.; Mravec, J.; Sakuragi, Y. Pectin Synthesis and Pollen Tube Growth in Arabidopsis Involves Three GAUT1 Golgi-Anchoring Proteins: GAUT5, GAUT6, and GAUT7. Front. Plant. Sci. 2020, 11, 585774. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Jensen, J.K.; Verhertbruggen, Y.; Søgaard, C.; Bernard, S.; Nafisi, M.; Poulsen, C.P.; Geshi, N.; Sakuragi, Y.; Driouich, A.; et al. ARAD Proteins Associated with Pectic Arabinan Biosynthesis Form Complexes When Transiently Overexpressed in Planta. Planta 2012, 236, 115–128. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Ho, Y.Y.; Moller, I.E.; Koh, P.L.; Golz, J.F.; Bacic, A.; Newbigin, E. A Glycosyltransferase from Nicotiana Alata Pollen Mediates Synthesis of a Linear (1,5)-α-L-Arabinan When Expressed in Arabidopsis. Plant Physiol. 2016, 170, 1962–1974. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Poulsen, P.C.; Vereb, G.; Kaneko, S.; Schulz, A.; Geshi, N. Galactosyltransferases from Arabidopsis Thaliana in the Biosynthesis of Type II Arabinogalactan: Molecular Interaction Enhances Enzyme Activity. BMC Plant Biol. 2014, 14, 90. [Google Scholar] [CrossRef]
- Schoberer, J.; Liebminger, E.; Botchway, S.W.; Strasser, R.; Hawes, C. Time-Resolved Fluorescence Imaging Reveals Differential Interactions of N-Glycan Processing Enzymes across the Golgi Stack in Planta. Plant Physiol. 2013, 161, 1737–1754. [Google Scholar] [CrossRef] [PubMed]
- Schoberer, J.; Izadi, S.; Kierein, C.; Vavra, U.; König-Beihammer, J.; Ruocco, V.; Grünwald-Gruber, C.; Castilho, A.; Strasser, R. The Tobacco GNTI Stem Region Harbors a Strong Motif for Homomeric Protein Complex Formation. Front. Plant Sci. 2023, 14, 1320051. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Jiang, N.; Nadella, R.; Killen, T.L.; Nadella, V.; Faik, A. A Glucurono (Arabino) Xylan Synthase Complex from Wheat Contains Members of the GT43, GT47, and GT75 Families and Functions Cooperatively. Plant Physiol. 2010, 154, 78–97. [Google Scholar] [CrossRef]
- Doong, R.L.; Mohnen, D. Solubilization and Characterization of a Galacturonosyltransferase that Synthesizes the Pectic Polysaccharide Homogalacturonan. Plant J. 1998, 13, 363–374. [Google Scholar] [CrossRef]
- Amos, R.A.; Pattathil, S.; Yang, J.Y.; Atmodjo, M.A.; Urbanowicz, B.R.; Moremen, K.W.; Mohnen, D. A Two-Phase Model for the Non-Processive Biosynthesis of Homogalacturonan Polysaccharides by the GAUT1:GAUT7 Complex. J. Biol. Chem. 2018, 293, 19047–19063. [Google Scholar] [CrossRef] [PubMed]
- Cicéron, F.; Rocha, J.; Kousar, S.; Hansen, S.F.; Chazalet, V.; Gillon, E.; Breton, C.; Lerouxel, O. Expression, Purification and Biochemical Characterization of AtFUT1, a Xyloglucan-Specific Fucosyltransferase from Arabidopsis Thaliana. Biochimie 2016, 128, 183–192. [Google Scholar] [CrossRef]
- Purushotham, P.; Ho, R.; Yu, L.; Fincher, G.B.; Bulone, V.; Zimmer, J. Mechanism of Mixed-Linkage Glucan Biosynthesis by Barley Cellulose Synthase-like CslF6 (1,3;1,4)-β-Glucan Synthase. Sci. Adv. 2022, 8, eadd1596. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Pereira, J.H.; Taujale, R.; Shao, W.; Bharadwaj, V.S.; Chapla, D.; Yang, J.Y.; Bomble, Y.J.; Moremen, K.W.; Kannan, N.; et al. Structural and Biochemical Insight into a Modular β-1,4-Galactan Synthase in Plants. Nat. Plants 2023, 9, 486–500. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zabotina, O.A. Critical Determinants in ER-Golgi Trafficking of Enzymes Involved in Glycosylation. Plants 2022, 11, 428. [Google Scholar] [CrossRef] [PubMed]
- Schoberer, J.; Vavra, U.; Stadlmann, J.; Hawes, C.; Mach, L.; Steinkellner, H.; Strasser, R. Arginine/Lysine Residues in the Cytoplasmic Tail Promote ER Export of Plant Glycosylation Enzyme. Traffic 2009, 10, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Schoberer, J.; Liebminger, E.; Vavra, U.; Veit, C.; Grünwald-Gruber, C.; Altmann, F.; Botchway, S.W.; Strasser, R. The Golgi Localization of Gnti Requires a Polar Amino Acid Residue within Its Transmembrane Domain. Plant Physiol. 2019, 180, 859–873. [Google Scholar] [CrossRef]
- Tu, L.; Tai, W.C.S.; Chen, L.; Banfield, D.K. Signal-Mediated Dynamic Retention of Glycosyltransferases in the Golgi. Science 2008, 321, 404–407. [Google Scholar] [CrossRef]
- Eckert, E.S.P.; Reckmann, I.; Hellwig, A.; Röhling, S.; El-Battari, A.; Wieland, F.T.; Popoff, V. Golgi Phosphoprotein 3 Triggers Signal-Mediated Incorporation of Glycosyltransferases into Coatomer-Coated (COPI) Vesicles. J. Biol. Chem. 2014, 289, 31319–31329. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Russo, D.; Kurokawa, K.; Sahu, P.; Lombardi, B.; Supino, D.; Zhukovsky, M.A.; Vocat, A.; Pothukuchi, P.; Kunnathully, V.; et al. Golgi Maturation-dependent Glycoenzyme Recycling Controls Glycosphingolipid Biosynthesis and Cell Growth via GOLPH3. EMBO J. 2021, 40, e107238. [Google Scholar] [CrossRef] [PubMed]
- Welch, L.G.; Peak-Chew, S.Y.; Begum, F.; Stevens, T.J.; Munro, S. Golph3 and Golph3l Are Broad-Spectrum Copi Adaptors for Sorting into Intra-Golgi Transport Vesicles. J. Cell Biol. 2021, 220, e202106115. [Google Scholar] [CrossRef]
- Liu, L.; Doray, B.; Kornfeld, S. Recycling of Golgi Glycosyltransferases Requires Direct Binding to Coatomer. Proc. Natl. Acad. Sci. USA 2018, 115, 8984–8989. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, C.G.; Maccioni, H.J.F. Endoplasmic Reticulum Export of Glycosyltransferases Depends on Interaction of a Cytoplasmic Dibasic Motif with Sar1. Mol. Biol. Cell 2003, 14, 3753–3766. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, J.J.; Weerts, R.M.; Shome, S.; Culbertson, A.T.; Honzatko, R.B.; Jernigan, R.L.; Zabotina, O.A. Xyloglucan Xylosyltransferase 1 Displays Promiscuity Toward Donor Substrates During in Vitro Reactions. Plant Cell Physiol. 2021, 62, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Phillips, D.R.; Ye, Z.H. A Single Xyloglucan Xylosyltransferase Is Sufficient for Generation of the XXXG Xylosylation Pattern of Xyloglucan. Plant Cell Physiol. 2021, 62, 1589–1602. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Julian, J.D.; Zabotina, O.A. Multiprotein Complexes of Plant Glycosyltransferases Involved in Their Function and Trafficking. Plants 2025, 14, 350. https://doi.org/10.3390/plants14030350
Zhang N, Julian JD, Zabotina OA. Multiprotein Complexes of Plant Glycosyltransferases Involved in Their Function and Trafficking. Plants. 2025; 14(3):350. https://doi.org/10.3390/plants14030350
Chicago/Turabian StyleZhang, Ning, Jordan D. Julian, and Olga A. Zabotina. 2025. "Multiprotein Complexes of Plant Glycosyltransferases Involved in Their Function and Trafficking" Plants 14, no. 3: 350. https://doi.org/10.3390/plants14030350
APA StyleZhang, N., Julian, J. D., & Zabotina, O. A. (2025). Multiprotein Complexes of Plant Glycosyltransferases Involved in Their Function and Trafficking. Plants, 14(3), 350. https://doi.org/10.3390/plants14030350







