Deciphering of Genomic Loci Associated with Alkaline Tolerance in Soybean [Glycine max (L.) Merr.] by Genome-Wide Association Study
Abstract
:1. Introduction
2. Results
2.1. Alkaline Treatment of Soybean Germplasm
2.2. Phenotypic Analysis of Alkaline Tolerance-Related Traits
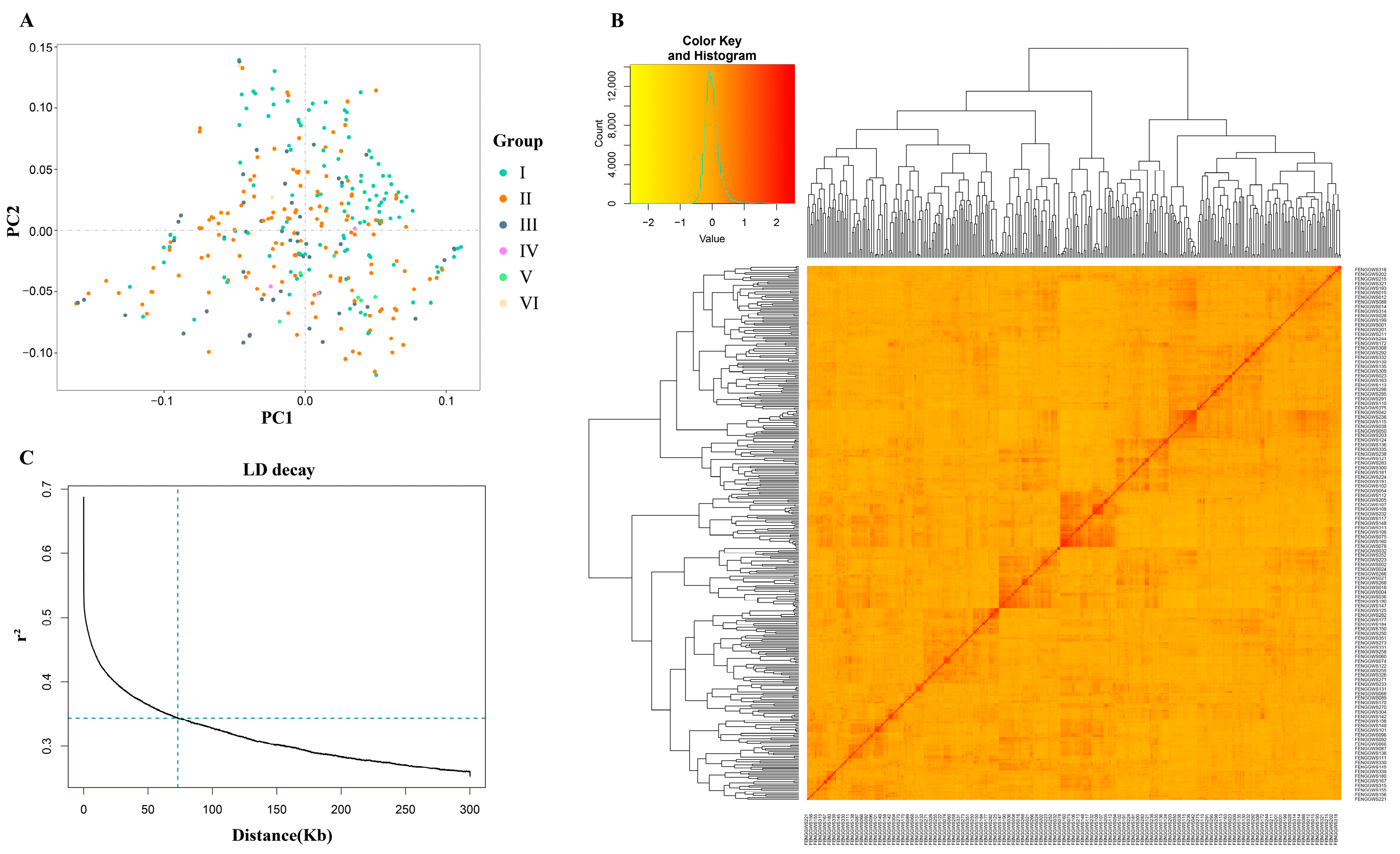
2.3. Population Structure and LD Analysis
2.4. Association Mapping Analysis of Alkaline Tolerance-Related Traits
2.5. Quantitative Trait Loci Analysis
2.6. Candidate Gene Identification
2.7. Haplotype Identification for Alkaline Tolerance
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Phenotypic Data Analysis
4.3. Genotyping, Population Structure, and Linkage Disequilibrium (LD) Analysis
4.4. Genome-Wide Association Study Analysis
4.5. Candidate Gene Analysis
4.6. Haplotype Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chaudhry, S.; Sidhu, G.P.S. Climate change regulated abiotic stress mechanisms in plants: A comprehensive review. Plant Cell Rep. 2022, 41, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Jat Baloch, M.Y.; Zhang, W.; Sultana, T.; Akram, M.; Shoumik, B.A.A.; Khan, M.Z.; Farooq, M.A. Utilization of sewage sludge to manage saline–alkali soil and increase crop production: Is it safe or not? Environ. Technol. Innov. 2023, 32, 103266. [Google Scholar] [CrossRef]
- Kothari, K.; Battisti, R.; Boote, K.J.; Archontoulis, S.V.; Confalone, A.; Constantin, J.; Cuadra, S.V.; Debaeke, P.; Faye, B.; Grant, B.; et al. Are soybean models ready for climate change food impact assessments? Eur. J. Agron. 2022, 135, 126482. [Google Scholar] [CrossRef]
- Tuyen, D.; Lal, S.; Xu, D. Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 2010, 121, 229–236. [Google Scholar]
- Qiao, Y.; Wang, Y.; Li, X.; Nisa, Z.U.; Jin, X.; Jing, L.; Yu, L.; Chen, C. Transcriptional profiling of alkaline stress-induced defense responses in soybean (Glycine max). Biotechnol. Biotec. Equip. 2021, 35, 1353–1360. [Google Scholar] [CrossRef]
- Wang, G.; Shen, W.; Zhang, Z.; Guo, S.; Hu, J.; Feng, R.; Zhao, Q.; Du, J.; Du, Y. The effect of neutral salt and alkaline stress with the same Na+ concentration on root growth of soybean (Glycine max (L.) Merr.) seedlings. Agronomy 2022, 12, 2708. [Google Scholar] [CrossRef]
- Cai, X.; Jia, B.; Sun, M.; Sun, X. Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant. Sci. 2022, 13, 1002302. [Google Scholar] [CrossRef]
- Bhat, J.A.; Yu, D. High-throughput NGS-based genotyping and phenotyping: Role in genomics-assisted breeding for soybean improvement. Legume Sci. 2021, 3, e81. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, X.; Jia, S.; Karikari, B.; Zhang, M.; Xia, Z.; Zhao, T.; Liang, F. Genome-wide association among soybean accessions for the genetic basis of salinity-alkalinity tolerance during germination. Crop Pasture Sci. 2021, 72, 255–267. [Google Scholar] [CrossRef]
- Zargar, S.M.; Raatz, B.; Sonah, H.; Nazir, M.; Bhat, J.A.; Dar, Z.A.; Agrawal, G.K.; Rakwal, R. Recent advances in molecular marker techniques: Insight into QTL mapping, GWAS and genomic selection in plants. J. Crop Sci. Biotechnol. 2015, 18, 293–308. [Google Scholar] [CrossRef]
- Do, T.D.; Vuong, T.D.; Dunn, D.; Clubb, M.; Valliyodan, B.; Patil, G.; Chen, P.; Xu, D.; Nguyen, H.T.; Shannon, J.G. Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genom. 2019, 20, 318. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37, 30. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Gou, Z.; Zhang, Y.; Wang, X.; Ren, H.; Wen, Z.; Kang, B.-K.; Li, Y.; Yu, L.; et al. Genome-wide association study of soybean seed germination under drought stress. Mol. Genet. Genom. 2020, 295, 661–673. [Google Scholar] [CrossRef]
- Yu, Z.; Chang, F.; Lv, W.; Sharmin, R.A.; Wang, Z.; Kong, J.; Bhat, J.A.; Zhao, T. Identification of QTN and candidate gene for seed-flooding tolerance in soybean [Glycine max (L.) Merr.] using genome-wide association study (GWAS). Genes 2019, 10, 957. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Han, D.; Yang, Q.; Li, C.; Shi, X.; Zhang, M.; Yang, C.; Qiu, L.; Jia, H.; et al. Cold tolerance SNPs and candidate gene mining in the soybean germination stage based on genome-wide association analysis. Theor. Appl. Genet. 2024, 137, 178. [Google Scholar] [CrossRef]
- Zong, C.; Zhao, J.; Wang, Y.; Wang, L.; Chen, Z.; Qi, Y.; Bai, Y.; Li, W.; Wang, W.; Ren, H.; et al. Identification of gene-allele system conferring alkali-tolerance at seedling stage in Northeast China soybean germplasm. Int. J. Mol. Sci. 2024, 25, 2963. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.; Baenziger, P.S.; Börner, A. GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from barley–a review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Kumar, V.; Vats, S.; Kumawat, S.; Bisht, A.; Bhatt, V.; Shivaraj, S.M.; Padalkar, G.; Goyal, V.; Zargar, S.; Gupta, S.; et al. Omics advances and integrative approaches for the simultaneous improvement of seed oil and protein content in soybean (Glycine max L.). Crit. Rev. Plant Sci. 2021, 40, 398–421. [Google Scholar] [CrossRef]
- Sagervanshi, A.; Geilfus, C.-M.; Kaiser, H.; Mühling, K.H. Alkali salt stress causes fast leaf apoplastic alkalinization together with shifts in ion and metabolite composition and transcription of key genes during the early adaptive response of Vicia faba L. Plant Sci. 2022, 319, 111253. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y. How do plants maintain pH and ion homeostasis under saline-alkali stress? Front. Plant Sci. 2023, 14, 1217193. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.H.; Liu, W.C.; Zhang, X.W.; Lu, Y.T. Ethylene inhibits root elongation during alkaline stress through AUXIN1 and associated changes in auxin accumulation. Plant Physiol. 2015, 168, 1777–1791. [Google Scholar] [CrossRef]
- Cao, Y.; Song, H.; Zhang, L. New insight into plant saline-alkali tolerance mechanisms and application to breeding. Int. J. Mol. Sci. 2022, 23, 16048. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Katagiri, T.; Iuchi, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Eggermont, L.; Verstraeten, B.; Van Damme, E.J.M. Genome-wide screening for lectin motifs in Arabidopsis thaliana. Plant Genome. 2017, 10, plantgenome2017.02.0010. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wei, R.; Xu, M.; Yao, W.; Jiang, J.; Ma, X.; Qu, G.; Jiang, T. Genome-wide analysis of HSF family and overexpression of PsnHSF21 confers salt tolerance in Populus simonii × P. nigra. Front. Plant Sci. 2023, 14, 1160102. [Google Scholar] [CrossRef]
- Song, Q.; He, F.; Kong, L.; Yang, J.; Wang, X.; Zhao, Z.; Zhang, Y.; Xu, C.; Fan, C.; Luo, K. The IAA17.1/HSFA5a module enhances salt tolerance in Populus tomentosa by regulating flavonol biosynthesis and ROS levels in lateral roots. New Phytol. 2024, 241, 592–606. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.; Yang, X.; Wu, J.; Cai, H.; Yang, N.; Li, X.; Wen, K.; Chen, D.; et al. Genome-wide identification and expression pattern analysis of the kiwifruit GRAS transcription factor family in response to salt stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 ligases and their role in abiotic stress response. Cells 2022, 11, 890. [Google Scholar] [CrossRef]
- Patil, G.; Do, T.; Vuong, T.D.; Valliyodan, B.; Lee, J.-D.; Chaudhary, J.; Shannon, J.G.; Nguyen, H.T. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 2016, 6, 19199. [Google Scholar] [CrossRef]
- Qian, L.; Hickey, L.T.; Stahl, A.; Werner, C.R.; Hayes, B.; Snowdon, R.J.; Voss-Fels, K.P. Exploring and harnessing haplotype diversity to improve yield stability in crops. Front. Plant Sci. 2017, 8, 1534. [Google Scholar] [CrossRef]
- Luján Basile, S.M.; Ramírez, I.A.; Crescente, J.M.; Conde, M.B.; Demichelis, M.; Abbate, P.; Rogers, W.J.; Pontaroli, A.C.; Helguera, M.; Vanzetti, L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019, 19, 553. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pang, Y.; Zhang, J.; Wu, Z.; Chen, K.; Ali, J.; Ye, G.; Xu, J.; Li, Z. Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci. Rep. 2017, 7, 17203. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Singh, V.K.; Saxena, R.K.; Khan, A.W.; Abbai, R.; Chitikineni, A.; Desai, A.; Molla, J.; Upadhyaya, H.D.; Kumar, A.; et al. Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan L.). Plant Biotechnol. J. 2020, 18, 2482–2490. [Google Scholar] [CrossRef]
- Zaitlen, N.A.; Kang, H.M.; Feolo, M.L.; Sherry, S.T.; Halperin, E.; Eskin, E. Inference and analysis of haplotypes from combined genotyping studies deposited in dbSNP. Genome Res. 2005, 15, 1594–1600. [Google Scholar] [CrossRef]
- Zhao, C.; Pan, X.; Yu, Y.; Zhu, Y.; Kong, F.; Sun, X.; Wang, F. Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breed. 2020, 40, 33. [Google Scholar] [CrossRef]
- Di, Q.; Dong, L.; Jiang, L.; Liu, X.; Cheng, P.; Liu, B.; Yu, G. Genome-wide association study and RNA-seq identifies GmWRI1-like transcription factor related to the seed weight in soybean. Front. Plant Sci. 2023, 14, 1268511. [Google Scholar] [CrossRef]
- Agoyi, E.E.; Ahomondji, S.E.; Yemadje, P.L.; Ayi, S.; Ranaivoson, L.; Torres, G.M.; da Fonseca Santos, M.; Boulakia, S.; Chigeza, G.; Assogbadjo, A.E.; et al. Combining AMMI and BLUP analysis to select high-yielding soybean genotypes in Benin. Agron. J. 2024, 116, 2109–2128. [Google Scholar] [CrossRef]
- El-Hashash, E. Genetic diversity of soybean yield based on cluster and principal component analyses. J. Adv. Biol. Biotechnol. 2016, 10, 1–9. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A SUPER powerful method for genome wide association study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Q.; Liu, Z.; Shi, X.; Wu, X.; Chen, Y.; Du, X.; Gao, Q.; He, D.; Shi, A.; et al. Genome-wide association analysis and genomic prediction of salt tolerance trait in soybean germplasm. Front. Plant Sci. 2024, 15, 1494551. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
- Cleophas, T.J.; Zwinderman, A.H.; Cleophas, T.F. Multiple Statistical Inferences, in Statistics Applied to Clinical Trials; Springer: Dordrecht, The Netherlands, 2006; pp. 87–96. [Google Scholar]




| Trait | Group | Min | Max | Median | Mean ± SD | CV% | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|
| RFW | CK | 0.89 | 7.32 | 3.16 | 3.35 ± 1.09 | 32.54 | 0.56 | 0.28 |
| AT | 0.15 | 3.47 | 1.28 | 1.36 ± 0.66 | 48.53 | 0.50 | −0.31 | |
| AT/CK | 0.04 | 1.33 | 0.40 | 0.42 ± 0.20 | 47.62 | 0.98 | 1.57 | |
| RDW | CK | 0.08 | 2.07 | 0.26 | 0.27 ± 0.13 | 48.15 | 7.98 | 106.01 |
| AT | 0.01 | 0.31 | 0.10 | 0.10 ± 0.06 | 60.00 | 0.57 | −0.31 | |
| AT/CK | 0.04 | 2.38 | 0.37 | 0.41 ± 0.26 | 63.41 | 2.24 | 10.48 | |
| SFW | CK | 1.56 | 12.06 | 5.77 | 5.90 ± 1.95 | 33.05 | 0.49 | −0.12 |
| AT | 0.24 | 3.43 | 1.03 | 1.26 ± 0.82 | 65.08 | 0.51 | −1.04 | |
| AT/CK | 0.04 | 1.04 | 0.18 | 0.22 ± 0.14 | 63.64 | 1.29 | 2.77 | |
| SDW | CK | 0.3 | 1.83 | 0.88 | 0.91 ± 0.30 | 32.97 | 0.58 | 0.13 |
| AT | 0.06 | 0.75 | 0.29 | 0.31 ± 0.12 | 38.71 | 0.71 | 0.04 | |
| AT/CK | 0.10 | 1.11 | 0.34 | 0.36 ± 0.15 | 41.67 | 1.34 | 3.33 | |
| RN | CK | 406.33 | 4269.33 | 2030.50 | 2108.10 ± 718.17 | 34.07 | 0.29 | −0.26 |
| AT | 79.00 | 1666.33 | 505.16 | 587.59 ± 335.61 | 57.12 | 0.78 | −0.08 | |
| AT/CK | 0.03 | 1.41 | 0.26 | 0.29 ± 0.17 | 58.62 | 1.86 | 6.58 | |
| RL | CK | 143.85 | 1457.82 | 734.00 | 742.17 ± 227.12 | 30.60 | 0.25 | −0.20 |
| AT | 41.37 | 621.99 | 226.25 | 248.98 ± 121.32 | 48.73 | 0.59 | −0.36 | |
| AT/CK | 0.04 | 1.52 | 0.32 | 0.35 ± 0.17 | 48.57 | 1.56 | 5.66 | |
| RTN | CK | 103.33 | 947.00 | 434.83 | 449.18 ± 150.85 | 33.58 | 0.65 | 0.43 |
| AT | 61.00 | 534.00 | 199.83 | 204.29 ± 77.06 | 37.72 | 0.55 | 0.33 | |
| AT/CK | 0.09 | 1.82 | 0.47 | 0.49 ± 0.22 | 44.90 | 1.35 | 4.54 | |
| CC | CK | 29.22 | 52.33 | 39.98 | 39.95 ± 3.87 | 9.69 | 0.11 | 0.19 |
| AT | 1.49 | 39.74 | 15.07 | 14.75 ± 10.17 | 68.95 | 0.20 | −1.32 | |
| AT/CK | 0.04 | 0.98 | 0.38 | 0.36 ± 0.24 | 66.67 | 0.19 | −1.30 |
| No. | Gene ID | Arabidopsis Ortholog | Gene Function Annotation |
|---|---|---|---|
| 1 | Glyma.01G113400 | AT4G00430 (plasma membrane intrinsic protein 1B) | Response to salt stress, response to temperature stimulus, response to water deprivation and water transport |
| 2 | Glyma.04G251900 | AT4G08250 (GRAS family transcription factor) | Regulation of transcription, DNA-dependent |
| 3 | Glyma.04G252100 | AT4G36020 (cold shock domain protein 1) | Response to cold, response to salt stress and response to water deprivation |
| 4 | Glyma.04G252300 | AT1G77690 (an auxin influx carrier LAX3) | Response to UV light, auxin polar transport, brassinosteroid biosynthetic process, response to auxin stimulus, response to cyclopentenone, root cap development and root hair elongation. |
| 5 | Glyma.04G252500 | AT4G08210 (Pentatricopeptide repeat (PPR-like) superfamily protein) | Biological process |
| 6 | Glyma.04G252600 | AT1G75710 (C2H2-like zinc finger protein) | NA |
| 7 | Glyma.04G252700 | AT1G77720 (PPK1, putative protein kinase 1) | DNA methylation, protein autophosphorylation, and protein phosphorylation |
| 8 | Glyma.04G253000 | AT4G08180 (OSBP (oxysterol binding protein)-related protein 1C) | Abscisic acid-mediated signaling pathway, response to cold, response to ethylene stimulus, and systemic acquired resistance |
| 9 | Glyma.04G253100 | AT1G21980 (PIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1) | Phosphatidylinositol metabolic process |
| 10 | Glyma.14G083700 | AT1G46264 (AtHSFB4, heat shock transcription factor B4) | Response to abiotic stress and response to heat |
| 11 | Glyma.14G083900 | AT1G45976 (S-ribonuclease binding protein 1) | Biological process; hormone-mediated signaling pathway; photoperiodism, flowering; signal transduction |
| 12 | Glyma.14G084500 | AT4G34110 (poly(A) binding protein 2) | Response to salt stress |
| 13 | Glyma.18G150300 | AT5G10530 (Concanavalin A-like lectin protein kinase family protein) | Protein phosphorylation |
| 14 | Glyma.20G072500 | AT5G55830 (LECRK-S.7, L-type lecting receptor kinase S.7) | Protein phosphorylation |
| 15 | Glyma.20G072600 | AT5G03540 (exocyst subunit exo70 family protein A1) | Auxin transport, hyperosmotic response, protein localization involved in auxin polar transport, response to salt stress, response to temperature stimulus, root development, and root hair elongation |
| 16 | Glyma.20G072700 | AT5G03540 (exocyst subunit exo70 family protein A1) | Auxin transport, hyperosmotic response, protein localization involved in auxin polar transport, response to salt stress, response to temperature stimulus, root development, and root hair elongation |
| 17 | Glyma.20G072900 | AT5G03540 (exocyst subunit exo70 family protein A1) | Auxin transport, hyperosmotic response, protein localization involved in auxin polar transport, response to salt stress, response to temperature stimulus, root development, and root hair elongation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Zhang, Y.; Bhat, J.A.; Wang, M.; Zheng, H.; Bu, M.; Zhao, B.; Yang, S.; Feng, X. Deciphering of Genomic Loci Associated with Alkaline Tolerance in Soybean [Glycine max (L.) Merr.] by Genome-Wide Association Study. Plants 2025, 14, 357. https://doi.org/10.3390/plants14030357
Yang X, Zhang Y, Bhat JA, Wang M, Zheng H, Bu M, Zhao B, Yang S, Feng X. Deciphering of Genomic Loci Associated with Alkaline Tolerance in Soybean [Glycine max (L.) Merr.] by Genome-Wide Association Study. Plants. 2025; 14(3):357. https://doi.org/10.3390/plants14030357
Chicago/Turabian StyleYang, Xinjing, Ye Zhang, Javaid Akhter Bhat, Mingjing Wang, Huanbin Zheng, Moran Bu, Beifang Zhao, Suxin Yang, and Xianzhong Feng. 2025. "Deciphering of Genomic Loci Associated with Alkaline Tolerance in Soybean [Glycine max (L.) Merr.] by Genome-Wide Association Study" Plants 14, no. 3: 357. https://doi.org/10.3390/plants14030357
APA StyleYang, X., Zhang, Y., Bhat, J. A., Wang, M., Zheng, H., Bu, M., Zhao, B., Yang, S., & Feng, X. (2025). Deciphering of Genomic Loci Associated with Alkaline Tolerance in Soybean [Glycine max (L.) Merr.] by Genome-Wide Association Study. Plants, 14(3), 357. https://doi.org/10.3390/plants14030357







