The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions
Abstract
1. Introduction
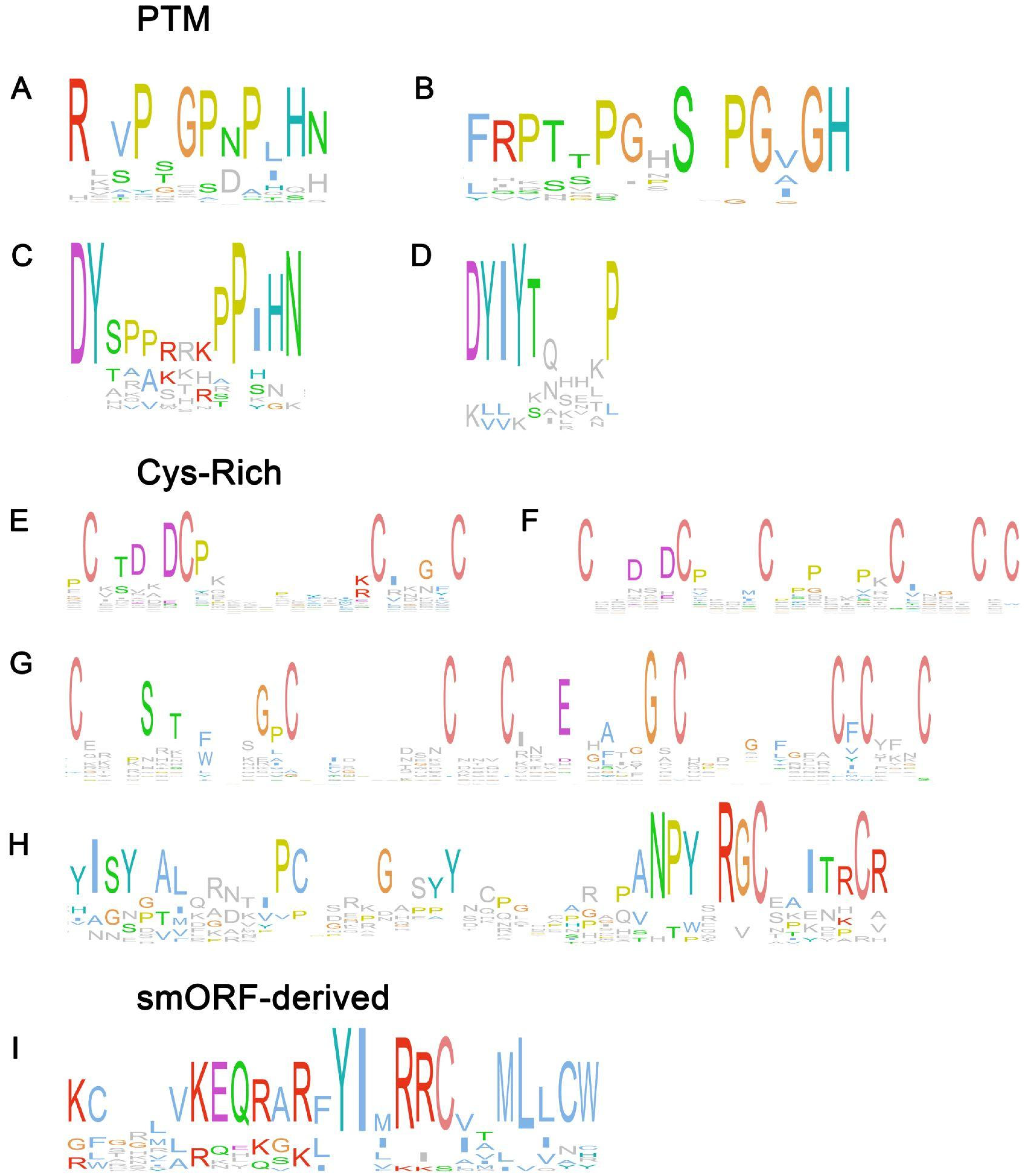
2. Peptide Families Involved in Symbiotic Interactions
2.1. PTM Peptide Families
2.1.1. CLE
2.1.2. CEP
2.1.3. RGF
2.1.4. PSK
2.2. Cys-Rich Peptide Families
2.2.1. NCR
2.2.2. DEF
2.2.3. RALF
2.2.4. nsLTP
2.3. Non-Cys Rich, Non-PTM Peptide Families
2.3.1. GRP
2.3.2. SNARP
2.4. SmORF-Derived Peptide Families
2.4.1. DVL
2.4.2. ENOD40
3. Peptides Involved in Both Symbiosis and Immunity
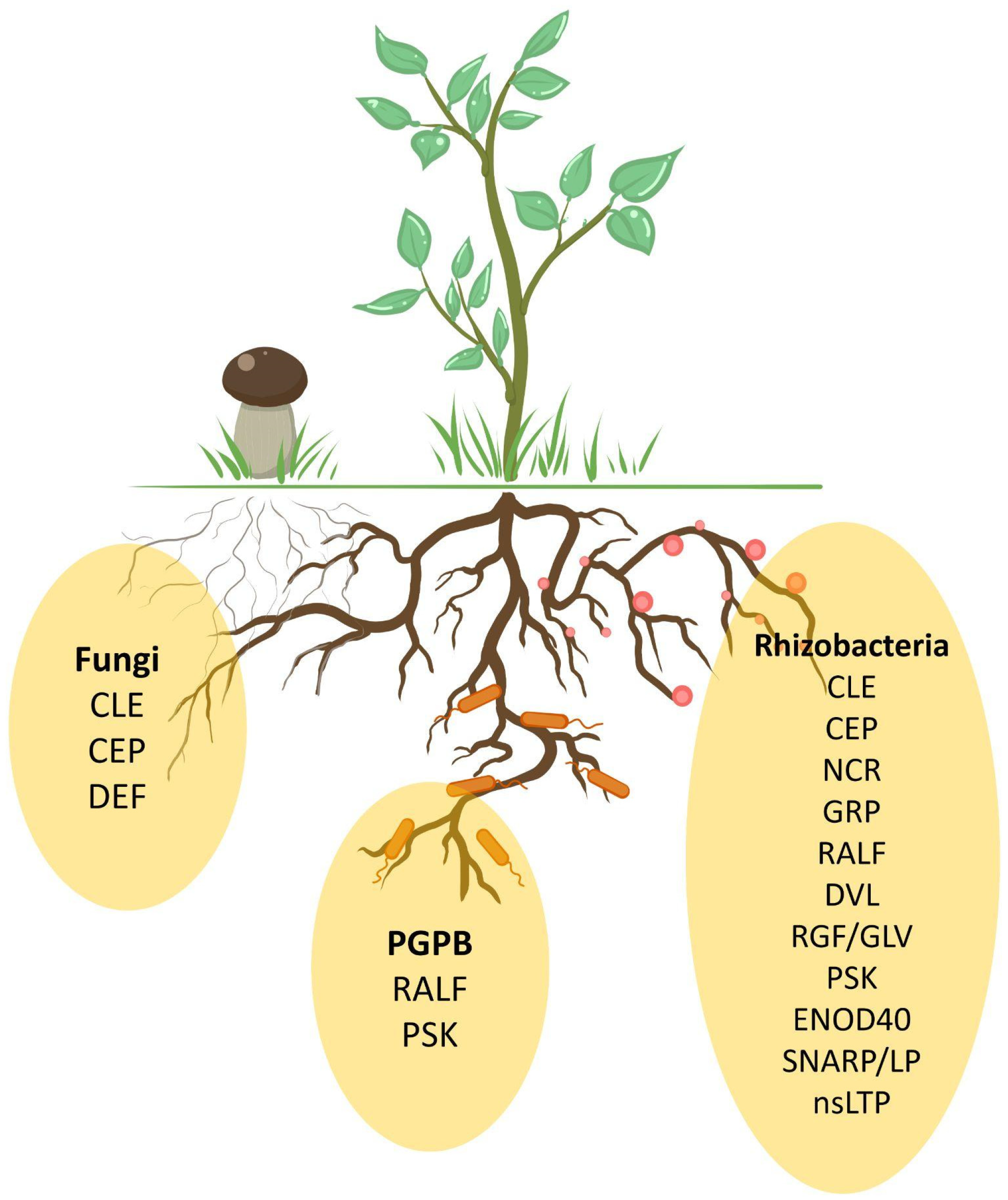
4. Rhizosphere Organisms Mimic Plant Peptides
5. Common Peptide Families for Different Symbiotic Interactions
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Delaux, P.-M.; Schornack, S. Plant Evolution Driven by Interactions with Symbiotic and Pathogenic Microbes. Science 2021, 371, eaba6605. [Google Scholar] [CrossRef]
- Middleton, H.; Yergeau, É.; Monard, C.; Combier, J.-P.; El Amrani, A. Rhizospheric Plant–Microbe Interactions: miRNAs as a Key Mediator. Trends Plant Sci. 2021, 26, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Varma, A. SymbiosSis: The Art of Living. In Symbiotic Fungi: Principles and Practice; Varma, A., Kharkwal, A.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–28. ISBN 978-3-540-95894-9. [Google Scholar]
- Lee, S.-M.; Ryu, C.-M. Algae as New Kids in the Beneficial Plant Microbiome. Front. Plant Sci. 2021, 12, 599742. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.; Lagunas, B.; Gifford, M.L. Determinants of Host Range Specificity in Legume-Rhizobia Symbiosis. Front. Microbiol. 2020, 11, 585749. [Google Scholar] [CrossRef]
- Downie, J.A.; Kondorosi, E. Why Should Nodule Cysteine-Rich (NCR) Peptides Be Absent From Nodules of Some Groups of Legumes but Essential for Symbiotic N-Fixation in Others? Front. Agron. 2021, 3, 654576. [Google Scholar] [CrossRef]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin Transport, Metabolism, and Signalling during Nodule Initiation: Indeterminate and Determinate Nodules. J. Exp. Bot. 2018, 69, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, P.; Uchiumi, T.; Alunni, B.; Evanno, G.; Cheron, A.; Catrice, O.; Mausset, A.-E.; Barloy-Hubler, F.; Galibert, F.; Kondorosi, A.; et al. Eukaryotic Control on Bacterial Cell Cycle and Differentiation in the Rhizobium–Legume Symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 5230–5235. [Google Scholar] [CrossRef]
- Pan, H.; Wang, D. Nodule Cysteine-Rich Peptides Maintain a Working Balance during Nitrogen-Fixing Symbiosis. Nat. Plants 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M. Developmental Biology of Legume Nodulation. New Phytol. 1992, 122, 211–237. [Google Scholar] [CrossRef] [PubMed]
- Froussart, E.; Bonneau, J.; Franche, C.; Bogusz, D. Recent Advances in Actinorhizal Symbiosis Signaling. Plant Mol. Biol. 2016, 90, 613–622. [Google Scholar] [CrossRef]
- Van Nguyen, T.; Pawlowski, K. Frankia and Actinorhizal Plants: Symbiotic Nitrogen Fixation. In Rhizotrophs: Plant Growth Promotion to Bioremediation; Mehnaz, S., Ed.; Springer: Singapore, 2017; pp. 237–261. ISBN 978-981-10-4862-3. [Google Scholar]
- Parniske, M. Arbuscular Mycorrhiza: The Mother of Plant Root Endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal Symbiosis in Plant Growth and Stress Adaptation: From Genes to Ecosystems. Annu. Rev. Plant Biol. 2023, 74, 569–607. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Minamisawa, K. Plant–Microbe Communications for Symbiosis. Plant Cell Physiol. 2010, 51, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.E.; Groten, K. The Costs and Benefits of Plant–Arbuscular Mycorrhizal Fungal Interactions. Annu. Rev. Plant Biol. 2022, 73, 649–672. [Google Scholar] [CrossRef]
- Hawkins, H.-J.; Cargill, R.I.M.; Van Nuland, M.E.; Hagen, S.C.; Field, K.J.; Sheldrake, M.; Soudzilovskaia, N.A.; Kiers, E.T. Mycorrhizal Mycelium as a Global Carbon Pool. Curr. Biol. 2023, 33, R560–R573. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Genre, A. Mechanisms Underlying Beneficial Plant–Fungus Interactions in Mycorrhizal Symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef]
- Mesny, F.; Hacquard, S.; Thomma, B.P. Co-evolution within the Plant Holobiont Drives Host Performance. EMBO Rep. 2023, 24, e57455. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.J.d.S.; Dias-Filho, M.B.; Gurgel, E.S.C. Successful Plant Growth-Promoting Microbes: Inoculation Methods and Abiotic Factors. Front. Sustain. Food Syst. 2021, 5, 606454. [Google Scholar] [CrossRef]
- Mergaert, P.; Kereszt, A.; Kondorosi, E. Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago Truncatula and Other Nodulating Plants. Plant Cell 2020, 32, 42–68. [Google Scholar] [CrossRef]
- Kang, W.; Jiang, Z.; Chen, Y.; Wu, F.; Liu, C.; Wang, H.; Shi, S.; Zhang, X.-X. Plant Transcriptome Analysis Reveals Specific Molecular Interactions between Alfalfa and Its Rhizobial Symbionts below the Species Level. BMC Plant Biol. 2020, 20, 293. [Google Scholar] [CrossRef]
- Kang, W.; Li, X.; Zhang, X.; Shi, S. Fine-Tuned Immune Antagonism and Nodule-Specific Cysteine-Rich Peptides Govern the Symbiotic Specificity Between Alfalfa Cultivars and Ensifer Meliloti. J. Plant Growth Regul. 2023, 42, 3696–3714. [Google Scholar] [CrossRef]
- Schnabel, E.L.; Chavan, S.A.; Gao, Y.; Poehlman, W.L.; Feltus, F.A.; Frugoli, J.A. A Medicago Truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2. Curr. Issues Mol. Biol. 2023, 45, 4612–4631. [Google Scholar] [CrossRef]
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-Seq Transcriptional Profiling of an Arbuscular Mycorrhiza Provides Insights into Regulated and Coordinated Gene Expression in Lotus Japonicus and Rhizophagus Irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef]
- Wheeler, J.I.; Irving, H.R. Plant Peptide Signaling: An Evolutionary Adaptation. In Plant Signaling Peptides; Irving, H.R., Gehring, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–23. ISBN 978-3-642-27603-3. [Google Scholar]
- Hu, X.-L.; Lu, H.; Hassan, M.M.; Zhang, J.; Yuan, G.; Abraham, P.E.; Shrestha, H.K.; Villalobos Solis, M.I.; Chen, J.-G.; Tschaplinski, T.J.; et al. Advances and Perspectives in Discovery and Functional Analysis of Small Secreted Proteins in Plants. Hortic Res 2021, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Pečenková, T.; Potocký, M.; Stegmann, M. More than Meets the Eye: Knowns and Unknowns of the Trafficking of Small Secreted Proteins in Arabidopsis. J. Exp. Bot. 2024, 75, 3713–3730. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P.A. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Malovichko, Y.V.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Plant Peptide Hormones. Russ. J. Plant Physiol. 2019, 66, 171–189. [Google Scholar] [CrossRef]
- Pandita, D.; Bhat, J.A.; Wani, S.H.; ElSayed, A.I.; Nawaz, G.; Mukherjee, S.; Reyes, V.P.; Kumar, A.; Shen, Q.; Ganie, S.A.; et al. Mobile Signaling Peptides: Secret Molecular Messengers with a Mighty Role in Plant Life. J. Plant Growth Regul. 2023, 42, 6801–6834. [Google Scholar] [CrossRef]
- Feng, Y.-Z.; Zhu, Q.-F.; Xue, J.; Chen, P.; Yu, Y. Shining in the Dark: The Big World of Small Peptides in Plants. aBIOTECH 2023, 4, 238–256. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tabata, R.; Sawa, S. Evolutionarily Conserved CLE Peptide Signaling in Plant Development, Symbiosis, and Parasitism. Curr. Opin. Plant Biol. 2013, 16, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Whitewoods, C. Evolution of CLE Peptide Signalling. Semin. Cell Dev. Biol. 2021, 109, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.L.; Ishida, T.; Sawa, S. CLE Peptides and Their Signaling Pathways in Plant Development. J. Exp. Bot. 2016, 67, 4813–4826. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Suzaki, T. Nitrate-Mediated Control of Root Nodule Symbiosis. Curr. Opin. Plant Biol. 2018, 44, 129–136. [Google Scholar] [CrossRef]
- Narasimhan, M.; Simon, R. Spatial Range, Temporal Span, and Promiscuity of CLE-RLK Signaling. Front Plant Sci 2022, 13, 906087. [Google Scholar] [CrossRef]
- Bashyal, S.; Gautam, C.K.; Müller, L.M. CLAVATA Signaling in Plant–Environment Interactions. Plant Physiol. 2024, 194, 1336–1357. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Lebedeva, M.; Dvornikova, K.; Lutova, L. Nitrate-Induced MtCLE34 Gene Lacks the Ability to Reduce Symbiotic Nodule Number and Carries Nonsense Mutation in a Few Accessions of Medicago Truncatula. Agronomy 2022, 12, 842. [Google Scholar] [CrossRef]
- Mens, C.; Hastwell, A.H.; Su, H.; Gresshoff, P.M.; Mathesius, U.; Ferguson, B.J. Characterisation of Medicago Truncatula CLE34 and CLE35 in Nitrate and Rhizobia Regulation of Nodulation. New Phytol. 2021, 229, 2525–2534. [Google Scholar] [CrossRef]
- Lebedeva, M.A.; Dobychkina, D.A.; Lutova, L.A. CRISPR/Cas9-Mediated Knock-Out of the MtCLE35 Gene Highlights Its Key Role in the Control of Symbiotic Nodule Numbers under High-Nitrate Conditions. Int. J. Mol. Sci. 2023, 24, 16816. [Google Scholar] [CrossRef] [PubMed]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’haeseleer, K.; Holsters, M.; Goormachtig, S. CLE Peptides Control Medicago Truncatula Nodulation Locally and Systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Handa, Y.; Tanaka, S.; Suzaki, T.; Kawaguchi, M. Expression of the CLE-RS3 Gene Suppresses Root Nodulation in Lotus Japonicus. J. Plant Res. 2016, 129, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.; Azarakhsh, M.; Yashenkova, Y.; Lutova, L. Nitrate-Induced CLE Peptide Systemically Inhibits Nodulation in Medicago Truncatula. Plants 2020, 9, 1456. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.; Dobychkina, D.; Yashenkova, Y.; Romanyuk, D.; Lutova, L. Local and Systemic Targets of the MtCLE35-SUNN Pathway in the Roots of Medicago truncatula. J. Plant Physiol. 2023, 281, 153922. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol. 2020, 30, 1339–1345.e3. [Google Scholar] [CrossRef]
- Valmas, M.I.; Sexauer, M.; Markmann, K.; Tsikou, D. Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis. Plants 2023, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.A.; Dobychkina, D.A.; Bashtovenko, K.A.; Petrenko, V.A.; Rubtsova, D.N.; Kochetkova, L.A.; Azarakhsh, M.; Romanyuk, D.A.; Lutova, L.A. MtCLE35 Mediates Inhibition of Rhizobia-Induced Signaling Pathway and Upregulation of Defense-Related Genes in Rhizobia-Inoculated Medicago Truncatula Roots. J. Plant Growth Regul. 2024, 43, 4941–4956. [Google Scholar] [CrossRef]
- Shinohara, H.; Matsubayashi, Y. Chemical Synthesis of Arabidopsis CLV3 Glycopeptide Reveals the Impact of Hydroxyproline Arabinosylation on Peptide Conformation and Activity. Plant Cell Physiol. 2013, 54, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-Derived CLE Glycopeptides Control Nodulation by Direct Binding to HAR1 Receptor Kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef]
- Imin, N.; Patel, N.; Corcilius, L.; Payne, R.J.; Djordjevic, M.A. CLE Peptide Tri-arabinosylation and Peptide Domain Sequence Composition Are Essential for SUNN-dependent Autoregulation of Nodulation in Medicago Truncatul. New Phytol. 2018, 218, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Yoro, E.; Suzaki, T.; Kawaguchi, M. CLE-HAR1 Systemic Signaling and NIN-Mediated Local Signaling Suppress the Increased Rhizobial Infection in the Daphne Mutant of Lotus Japonicus. MPMI 2020, 33, 320–327. [Google Scholar] [CrossRef]
- Kassaw, T.; Nowak, S.; Schnabel, E.; Frugoli, J. ROOT DETERMINED NODULATION1 Is Required for M. Truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol. 2017, 174, 2445–2456. [Google Scholar] [CrossRef] [PubMed]
- Karlo, M.; Boschiero, C.; Landerslev, K.G.; Blanco, G.S.; Wen, J.; Mysore, K.S.; Dai, X.; Zhao, P.X.; de Bang, T.C. The CLE53–SUNN Genetic Pathway Negatively Regulates Arbuscular Mycorrhiza Root Colonization in Medicago Truncatula. J. Exp. Bot. 2020, 71, 4972–4984. [Google Scholar] [CrossRef]
- Müller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE–SUNN Module Regulates Strigolactone Content and Fungal Colonization in Arbuscular Mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant Sesquiterpenes Induce Hyphal Branching in Arbuscular Mycorrhizal Fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Kodama, K.; Rich, M.K.; Yoda, A.; Shimazaki, S.; Xie, X.; Akiyama, K.; Mizuno, Y.; Komatsu, A.; Luo, Y.; Suzuki, H.; et al. An Ancestral Function of Strigolactones as Symbiotic Rhizosphere Signals. Nat. Commun. 2022, 13, 3974. [Google Scholar] [CrossRef] [PubMed]
- Wulf, K.; Sun, J.; Wang, C.; Ho-Plagaro, T.; Kwon, C.-T.; Velandia, K.; Correa-Lozano, A.; Tamayo-Navarrete, M.I.; Reid, J.B.; García Garrido, J.M.; et al. The Role of CLE Peptides in the Suppression of Mycorrhizal Colonization of Tomato. Plant Cell Physiol. 2024, 65, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Velandia, K.; Kwon, C.-T.; Wulf, K.E.; Nichols, D.S.; Reid, J.B.; Foo, E. The Role of CLAVATA Signalling in the Negative Regulation of Mycorrhizal Colonization and Nitrogen Response of Tomato. J. Exp. Bot. 2021, 72, 1702–1713. [Google Scholar] [CrossRef]
- Marquer, M.L.; Bécard, G.; Frey, N.F. dit Arbuscular Mycorrhizal Fungi Possess a CLAVATA3/Embryo Surrounding Region-related Gene That Positively Regulates Symbiosis. New Phytol. 2018, 222, 1030–1042. [Google Scholar] [CrossRef]
- Taleski, M.; Jin, M.; Chapman, K.; Taylor, K.; Winning, C.; Frank, M.; Imin, N.; Djordjevic, M.A. CEP Hormones at the Nexus of Nutrient Acquisition and Allocation, Root Development, and Plant–Microbe Interactions. J. Exp. Bot. 2024, 75, 538–552. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, H.A.; Imin, N.; Djordjevic, M.A. Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) Gene Family in Angiosperms, and Evolution of Plant-Family Specific CEP Genes. BMC Genom. 2014, 15, 870. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Frugier, F. Rhizobium Symbiotic Efficiency Meets CEP Signaling Peptides. New Phytol. 2023, 241, 24–27. [Google Scholar] [CrossRef]
- Lebedeva, M.A.; Gancheva, M.S.; Kulaeva, O.A.; Zorin, E.A.; Dobychkina, D.A.; Romanyuk, D.A.; Sulima, A.S.; Zhukov, V.A.; Lutova, L.A. Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L. Int. J. Mol. Sci. 2022, 23, 14875. [Google Scholar] [CrossRef] [PubMed]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The Peptide-Encoding CEP1 Gene Modulates Lateral Root and Nodule Numbers in Medicago Truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef]
- Wu, S.; Wang, X.; Qin, J.; Tian, W.; Wang, M.; Yue, A.; Wang, L.; Du, W.; Zhao, J. Soybean CEP6 Signaling Peptides Positively Regulate Nodulation. Agronomy 2024, 14, 988. [Google Scholar] [CrossRef]
- Zhu, F.; Deng, J.; Chen, H.; Liu, P.; Zheng, L.; Ye, Q.; Li, R.; Brault, M.; Wen, J.; Frugier, F.; et al. A CEP Peptide Receptor-Like Kinase Regulates Auxin Biosynthesis and Ethylene Signaling to Coordinate Root Growth and Symbiotic Nodulation in Medicago Truncatula. Plant Cell 2020, 32, 2855–2877. [Google Scholar] [CrossRef]
- Hsieh, Y.-H.; Wei, Y.-H.; Lo, J.-C.; Pan, H.-Y.; Yang, S.-Y. Arbuscular Mycorrhizal Symbiosis Enhances Tomato Lateral Root Formation by Modulating CEP2 Peptide Expression. New Phytol. 2022, 235, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Pedinotti, L.; Teyssendier de la Serve, J.; Roudaire, T.; San Clemente, H.; Aguilar, M.; Kohlen, W.; Frugier, F.; Frei dit Frey, N. The CEP Peptide-CRA2 Receptor Module Promotes Arbuscular Mycorrhizal Symbiosis. Curr. Biol. 2024, 4, 5366–5373.e4. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Müller, L.M. A Rulebook for Peptide Control of Legume–Microbe Endosymbioses. Trends Plant Sci. 2022, 27, 870–889. [Google Scholar] [CrossRef] [PubMed]
- Gautrat, P.; Laffont, C.; Frugier, F.; Ruffel, S. Nitrogen Systemic Signaling: From Symbiotic Nodulation to Root Acquisition. Trends Plant Sci. 2021, 26, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN Transcription Factor Coordinates CEP and CLE Signaling Peptides That Regulate Nodulation Antagonistically. Nat. Commun. 2020, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Drozdzecki, A.; Hoogewijs, K.; Nguyen, A.; Beeckman, T.; Madder, A.; Hilson, P. Transcriptional and Functional Classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-Like Signaling Peptides Reveals Their Role in Lateral Root and Hair Formation. Plant Physiol. 2013, 161, 954–970. [Google Scholar] [CrossRef]
- Li, Q.; Li, M.; Zhang, D.; Yu, L.; Yan, J.; Luo, L. The Peptide-Encoding MtRGF3 Gene Negatively Regulates Nodulation of Medicago truncatula. Biochem. Biophys. Res. Commun. 2020, 523, 66–71. [Google Scholar] [CrossRef]
- Roy, S.; Torres-Jerez, I.; Zhang, S.; Liu, W.; Schiessl, K.; Jain, D.; Boschiero, C.; Lee, H.-K.; Krom, N.; Zhao, P.X.; et al. The Peptide GOLVEN10 Alters Root Development and Noduletaxis in Medicago Truncatula. Plant J. 2024, 118, 607–625. [Google Scholar] [CrossRef]
- Li, Q.; Shan, D.; Zheng, W.; Wang, Y.; Lin, Z.; Jin, H.; Ding, A.; Yan, J.; Yu, L.; Luo, L. MtRGF3 Peptide Activates Defense Responses and Represses the Expressions of Nodulation Signaling Genes in Medicago Truncatula. ABBS 2023, 55, 1319–1322. [Google Scholar] [CrossRef]
- Stegmann, M.; Zecua-Ramirez, P.; Ludwig, C.; Lee, H.-S.; Peterson, B.; Nimchuk, Z.L.; Belkhadir, Y.; Hückelhoven, R. RGI-GOLVEN Signaling Promotes Cell Surface Immune Receptor Abundance to Regulate Plant Immunity. EMBO Rep. 2022, 23, e53281. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Di, Q.; Luo, L.; Yu, L. Phytosulfokine Peptides, Their Receptors, and Functions. Front. Plant Sci. 2024, 14, 1326964. [Google Scholar] [CrossRef]
- Shen, X.; Stührwohldt, N.; Lin, C. The Research Process of PSK Biosynthesis, Signaling Transduction, and Potential Applications in Brassica Napus. Plants 2023, 12, 3075. [Google Scholar] [CrossRef]
- Wang, C.; Yu, H.; Zhang, Z.; Yu, L.; Xu, X.; Hong, Z.; Luo, L. Phytosulfokine Is Involved in Positive Regulation of Lotus Japonicus Nodulation. MPMI 2015, 28, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Di, Q.; Zhang, D.; Liu, Y.; Li, X.; Mysore, K.S.; Wen, J.; Yan, J.; Luo, L. A Legume-Specific Novel Type of Phytosulfokine, PSK-δ, Promotes Nodulation by Enhancing Nodule Organogenesis. J. Exp. Bot. 2022, 73, 2698–2713. [Google Scholar] [CrossRef]
- Di, Q.; Li, Y.; Zhang, D.; Wu, W.; Zhang, L.; Zhao, X.; Luo, L.; Yu, L. A Novel Type of Phytosulfokine, PSK-ε, Positively Regulates Root Elongation and Formation of Lateral Roots and Root Nodules in Medicago Truncatula. Plant Signal. Behav. 2022, 17, 2134672. [Google Scholar] [CrossRef]
- Suri, G.S.; Tiwari, M. Phytosulfokine-δ: A Small Peptide, but a Big Player in Symbiosis Gene Regulation. Int. J. Plant Biol. 2023, 14, 100–103. [Google Scholar] [CrossRef]
- Song, S.; Morales Moreira, Z.; Briggs, A.L.; Zhang, X.-C.; Diener, A.C.; Haney, C.H. PSKR1 Balances the Plant Growth–Defence Trade-off in the Rhizosphere Microbiome. Nat. Plants 2023, 9, 2071–2084. [Google Scholar] [CrossRef]
- Tan, W.; Nian, H.; Tran, L.-S.P.; Jin, J.; Lian, T. Small Peptides: Novel Targets for Modulating Plant–Rhizosphere Microbe Interactions. Trends Microbiol. 2024, 32, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.M.; Kylarová, S.; Mergaert, P.; Kondorosi, É. Unexplored Arsenals of Legume Peptides With Potential for Their Applications in Medicine and Agriculture. Front. Microbiol. 2020, 11, 1307. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.; Shirdel, I.; Braun, M.S.; Wink, M. Defensins: Transcriptional Regulation and Function beyond Antimicrobial Activity. Dev. Comp. Immunol. 2020, 104, 103556. [Google Scholar] [CrossRef]
- Salgado, M.G.; Demina, I.V.; Maity, P.J.; Nagchowdhury, A.; Caputo, A.; Krol, E.; Loderer, C.; Muth, G.; Becker, A.; Pawlowski, K. Legume NCRs and Nodule-Specific Defensins of Actinorhizal Plants—Do They Share a Common Origin? PLoS ONE 2022, 17, e0268683. [Google Scholar] [CrossRef]
- Hanks, J.N.; Snyder, A.K.; Graham, M.A.; Shah, R.K.; Blaylock, L.A.; Harrison, M.J.; Shah, D.M. Defensin Gene Family in Medicago Truncatula: Structure, Expression and Induction by Signal Molecules. Plant Mol. Biol. 2005, 58, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Uhe, M.; Hogekamp, C.; Hartmann, R.M.; Hohnjec, N.; Küster, H. The Mycorrhiza-Dependent Defensin MtDefMd1 of Medicago Truncatula Acts during the Late Restructuring Stages of Arbuscule-Containing Cells. PLoS ONE 2018, 13, e0191841. [Google Scholar] [CrossRef] [PubMed]
- Moola, A.K.; Gurusamy, D.; Arya, S.K.; Sivakumar, J.S.; Elavarasan, K.; Vasanth, K.; Balasubramani, S. Chapter 10—Defensins in Plants: Diversity and Role in Plant Defense. In Defense-Related Proteins in Plants; Upadhyay, S.K., Ed.; Academic Press: Cambridge, MA, USA, 2024; pp. 263–281. ISBN 978-0-443-13236-0. [Google Scholar]
- Isozumi, N.; Masubuchi, Y.; Imamura, T.; Mori, M.; Koga, H.; Ohki, S. Structure and Antimicrobial Activity of NCR169, a Nodule-Specific Cysteine-Rich Peptide of Medicago Truncatula. Sci. Rep. 2021, 11, 9923. [Google Scholar] [CrossRef]
- Horváth, B.; Güngör, B.; Tóth, M.; Domonkos, Á.; Ayaydin, F.; Saifi, F.; Chen, Y.; Biró, J.B.; Bourge, M.; Szabó, Z.; et al. The Medicago Truncatula Nodule-Specific Cysteine-Rich Peptides, NCR343 and NCR-New35 Are Required for the Maintenance of Rhizobia in Nitrogen-Fixing Nodules. New Phytol. 2023, 239, 1974–1988. [Google Scholar] [CrossRef]
- Zhang, R.; Shen, Y.; He, J.; Zhang, C.; Ma, Y.; Sun, C.; Song, X.; Li, L.; Zhang, S.; Biró, J.B.; et al. Nodule-Specific Cysteine-Rich Peptide 343 Is Required for Symbiotic Nitrogen Fixation in Medicago Truncatula. Plant Physiol. 2023, 193, 1897–1912. [Google Scholar] [CrossRef]
- Kim, M.; Chen, Y.; Xi, J.; Waters, C.; Chen, R.; Wang, D. An Antimicrobial Peptide Essential for Bacterial Survival in the Nitrogen-Fixing Symbiosis. Proc. Natl. Acad. Sci. USA 2015, 112, 15238–15243. [Google Scholar] [CrossRef] [PubMed]
- Jenei, S.; Tiricz, H.; Szolomájer, J.; Tímár, E.; Klement, É.; Al Bouni, M.A.; Lima, R.M.; Kata, D.; Harmati, M.; Buzás, K.; et al. Potent Chimeric Antimicrobial Derivatives of the Medicago Truncatula NCR247 Symbiotic Peptide. Front. Microbiol. 2020, 11, 270. [Google Scholar] [CrossRef]
- Howan, D.H.O.; Jenei, S.; Szolomajer, J.; Endre, G.; Kondorosi, É.; Tóth, G.K. Enhanced Antibacterial Activity of Substituted Derivatives of NCR169C Peptide. Int. J. Mol. Sci. 2023, 24, 2694. [Google Scholar] [CrossRef]
- Montiel, J.; Downie, J.A.; Farkas, A.; Bihari, P.; Herczeg, R.; Bálint, B.; Mergaert, P.; Kereszt, A.; Kondorosi, É. Morphotype of Bacteroids in Different Legumes Correlates with the Number and Type of Symbiotic NCR Peptides. Proc. Natl. Acad. Sci. USA 2017, 114, 5041–5046. [Google Scholar] [CrossRef]
- Saifi, F.; Biró, J.B.; Horváth, B.; Vizler, C.; Laczi, K.; Rákhely, G.; Kovács, S.; Kang, M.; Li, D.; Chen, Y.; et al. Two Members of a Nodule-specific Cysteine-Rich (NCR) Peptide Gene Cluster Are Required for Differentiation of Rhizobia in Medicago Truncatula Nodules. Plant J. 2024, 119, 1508–1525. [Google Scholar] [CrossRef] [PubMed]
- Horváth, B.; Domonkos, Á.; Kereszt, A.; Szűcs, A.; Ábrahám, E.; Ayaydin, F.; Bóka, K.; Chen, Y.; Chen, R.; Murray, J.D.; et al. Loss of the Nodule-Specific Cysteine Rich Peptide, NCR169, Abolishes Symbiotic Nitrogen Fixation in the Medicago Truncatula Dnf7 Mutant. Proc. Natl. Acad. Sci. USA 2015, 112, 15232–15237. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Q.; Fedorova, E.; Liu, J.; Qin, Q.; Zheng, Q.; Price, P.A.; Pan, H.; Wang, D.; Griffitts, J.S.; et al. Microsymbiont Discrimination Mediated by a Host-Secreted Peptide in Medicago Truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6848–6853. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Ábrahám, E.; Gombár, A.; Domonkos, Á.; Szűcs, A.; Körmöczi, P.; Wang, T.; et al. Host-Secreted Antimicrobial Peptide Enforces Symbiotic Selectivity in Medicago Truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Liu, Y.; Zhou, D.; Zhao, W.; Chen, Z.; Chen, D.; Li, Y.; Zhang, X.-X. Transcriptomic Identification of a Unique Set of Nodule-Specific Cysteine-Rich Peptides Expressed in the Nitrogen-Fixing Root Nodule of Astragalus Sinicus. MPMI 2022, 35, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Güngör, B.; Biró, J.B.; Domonkos, Á.; Horváth, B.; Kaló, P. Targeted Mutagenesis of Medicago Truncatula Nodule-Specific Cysteine-Rich (NCR) Genes Using the Agrobacterium Rhizogenes-Mediated CRISPR/Cas9 System. Sci. Rep. 2023, 13, 20676. [Google Scholar] [CrossRef]
- Shabab, M.; Arnold, M.F.F.; Penterman, J.; Wommack, A.J.; Bocker, H.T.; Price, P.A.; Griffitts, J.S.; Nolan, E.M.; Walker, G.C. Disulfide Cross-Linking Influences Symbiotic Activities of Nodule Peptide NCR247. Proc. Natl. Acad. Sci. USA 2016, 113, 10157–10162. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.W.; Baldacci-Cresp, F.; Pierre, O.; Larousse, M.; Benyamina, S.; Lambert, A.; Hopkins, J.; Castella, C.; Cazareth, J.; Alloing, G.; et al. Regulation of Differentiation of Nitrogen-Fixing Bacteria by Microsymbiont Targeting of Plant Thioredoxin S1. Curr. Biol. 2017, 27, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant Peptides Govern Terminal Differentiation of Bacteria in Symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Benedict, A.B.; Ghosh, P.; Scott, S.M.; Griffitts, J.S. A Conserved Rhizobial Peptidase That Interacts with Host-Derived Symbiotic Peptides. Sci. Rep. 2021, 11, 11779. [Google Scholar] [CrossRef] [PubMed]
- Wendlandt, C.E.; Helliwell, E.; Roberts, M.; Nguyen, K.T.; Friesen, M.L.; von Wettberg, E.; Price, P.; Griffitts, J.S.; Porter, S.S. Decreased Coevolutionary Potential and Increased Symbiont Fecundity during the Biological Invasion of a Legume-Rhizobium Mutualism. Evolution 2021, 75, 731–747. [Google Scholar] [CrossRef]
- Price, P.A.; Tanner, H.R.; Dillon, B.A.; Shabab, M.; Walker, G.C.; Griffitts, J.S. Rhizobial Peptidase HrrP Cleaves Host-Encoded Signaling Peptides and Mediates Symbiotic Compatibility. Proc. Natl. Acad. Sci. USA 2015, 112, 15244–15249. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Chakraborty, J. Exploring the Role of Symbiotic Modifier Peptidases in the Legume − Rhizobium Symbiosis. Arch. Microbiol. 2024, 206, 147. [Google Scholar] [CrossRef] [PubMed]
- Penterman, J.; Abo, R.P.; De Nisco, N.J.; Arnold, M.F.F.; Longhi, R.; Zanda, M.; Walker, G.C. Host Plant Peptides Elicit a Transcriptional Response to Control the Sinorhizobium Meliloti Cell Cycle during Symbiosis. Proc. Natl. Acad. Sci. USA 2014, 111, 3561–3566. [Google Scholar] [CrossRef]
- Sankari, S.; Babu, V.M.P.; Bian, K.; Alhhazmi, A.; Andorfer, M.C.; Avalos, D.M.; Smith, T.A.; Yoon, K.; Drennan, C.L.; Yaffe, M.B.; et al. A Haem-Sequestering Plant Peptide Promotes Iron Uptake in Symbiotic Bacteria. Nat. Microbiol. 2022, 7, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Quilbé, J.; Lamy, L.; Brottier, L.; Leleux, P.; Fardoux, J.; Rivallan, R.; Benichou, T.; Guyonnet, R.; Becana, M.; Villar, I.; et al. Genetics of Nodulation in Aeschynomene Evenia Uncovers Mechanisms of the Rhizobium–Legume Symbiosis. Nat. Commun. 2021, 12, 829. [Google Scholar] [CrossRef] [PubMed]
- Libourel, C.; Keller, J.; Brichet, L.; Cazalé, A.-C.; Carrère, S.; Vernié, T.; Couzigou, J.-M.; Callot, C.; Dufau, I.; Cauet, S.; et al. Comparative Phylotranscriptomics Reveals Ancestral and Derived Root Nodule Symbiosis Programmes. Nat. Plants 2023, 9, 1067–1080. [Google Scholar] [CrossRef]
- Kaur, J.; Fellers, J.; Adholeya, A.; Velivelli, S.L.S.; El-Mounadi, K.; Nersesian, N.; Clemente, T.; Shah, D. Expression of Apoplast-Targeted Plant Defensin MtDef4.2 Confers Resistance to Leaf Rust Pathogen Puccinia Triticina but Does Not Affect Mycorrhizal Symbiosis in Transgenic Wheat. Transgenic Res. 2017, 26, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; De Smet, I. Understanding the RALF Family: A Tale of Many Species. Trends Plant Sci. 2014, 19, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Tena, G. Immunity: RALF to the Rescue. Nat. Plants 2016, 2, 16067. [Google Scholar] [CrossRef]
- Wieghaus, A.; Prüfer, D.; Gronover, C.S. Loss of Function Mutation of the Rapid Alkalinization Factor (RALF1)-like Peptide in the Dandelion Taraxacum Koksaghyz Entails a High-Biomass Taproot Phenotype. PLoS ONE 2019, 14, e0217454. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The Receptor Kinase FER Is a RALF-Regulated Scaffold Controlling Plant Immune Signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Mamaeva, A.; Lyapina, I.; Knyazev, A.; Golub, N.; Mollaev, T.; Chudinova, E.; Elansky, S.; Babenko, V.V.; Veselovsky, V.A.; Klimina, K.M.; et al. RALF Peptides Modulate Immune Response in the Moss Physcomitrium Patens. Front. Plant Sci. 2023, 14, 1077301. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, X.; Bai, Z.; Tanveer, M.; Zhang, Y.; Chen, W.; Shabala, S.; Huang, L. Small but Powerful: RALF Peptides in Plant Adaptive and Developmental Responses. Plant Sci. 2024, 343, 112085. [Google Scholar] [CrossRef]
- Campbell, L.; Turner, S.R. A Comprehensive Analysis of RALF Proteins in Green Plants Suggests There Are Two Distinct Functional Groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Solís-Miranda, J.; Juárez-Verdayes, M.A.; Nava, N.; Rosas, P.; Leija-Salas, A.; Cárdenas, L.; Quinto, C. The Phaseolus Vulgaris Receptor-Like Kinase PvFER1 and the Small Peptides PvRALF1 and PvRALF6 Regulate Nodule Number as a Function of Nitrate Availability. Int. J. Mol. Sci. 2023, 24, 5230. [Google Scholar] [CrossRef]
- Combier, J.-P.; Küster, H.; Journet, E.-P.; Hohnjec, N.; Gamas, P.; Niebel, A. Evidence for the Involvement in Nodulation of the Two Small Putative Regulatory Peptide-Encoding Genes MtRALFL1 and MtDVL1. MPMI 2008, 21, 1118–1127. [Google Scholar] [CrossRef]
- Jia, Y.; Li, Y. Genome-Wide Identification and Comparative Analysis of RALF Gene Family in Legume and Non-Legume Species. Int. J. Mol. Sci. 2023, 24, 8842. [Google Scholar] [CrossRef]
- Song, Y.; Wilson, A.J.; Zhang, X.-C.; Thoms, D.; Sohrabi, R.; Song, S.; Geissmann, Q.; Liu, Y.; Walgren, L.; He, S.Y.; et al. FERONIA Restricts Pseudomonas in the Rhizosphere Microbiome via Regulation of Reactive Oxygen Species. Nat. Plants 2021, 7, 644–654. [Google Scholar] [CrossRef]
- Tang, J.; Wu, D.; Li, X.; Wang, L.; Xu, L.; Zhang, Y.; Xu, F.; Liu, H.; Xie, Q.; Dai, S.; et al. Plant Immunity Suppression via PHR1-RALF-FERONIA Shapes the Root Microbiome to Alleviate Phosphate Starvation. EMBO J. 2022, 41, e109102. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Morea, F.A.; Liu, J.; Shan, L.; He, P. Malectin-like Receptor Kinases as Protector Deities in Plant Immunity. Nat. Plants 2022, 8, 27–37. [Google Scholar] [CrossRef]
- Liao, Y.; Wen, X.; Zheng, J.; Li, X.; Deng, X.; Tan, X.; Liang, X.; Yi, X.; Liao, H. RALF-like Peptide Improves the Colonization of Endophytic Colletotrichum Tofieldiae through Interacting with Plant Receptor-like Kinase. Plant Pathol. 2023, 72, 1649–1661. [Google Scholar] [CrossRef]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G.; et al. A Fungal Pathogen Secretes Plant Alkalinizing Peptides to Increase Infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Peng, H.; Zhu, S.; Xing, J.; Li, X.; Zhu, Z.; Zheng, J.; Wang, L.; Wang, B.; Chen, J.; et al. Nematode-Encoded RALF Peptide Mimics Facilitate Parasitism of Plants through the FERONIA Receptor Kinase. Mol. Plant 2020, 13, 1434–1454. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-García, C.; Solis-Miranda, J.; Pacheco, R.; Quinto, C. Non-Specific Lipid Transfer Proteins in Legumes and Their Participation During Root-Nodule Symbiosis. Front. Agron. 2021, 3, 660100. [Google Scholar] [CrossRef]
- Missaoui, K.; Gonzalez-Klein, Z.; Pazos-Castro, D.; Hernandez-Ramirez, G.; Garrido-Arandia, M.; Brini, F.; Diaz-Perales, A.; Tome-Amat, J. Plant Non-Specific Lipid Transfer Proteins: An Overview. Plant Physiol. Biochem. 2022, 171, 115–127. [Google Scholar] [CrossRef]
- Gao, S.; Guo, W.; Feng, W.; Liu, L.; Song, X.; Chen, J.; Hou, W.; Zhu, H.; Tang, S.; Hu, J. LTP3 Contributes to Disease Susceptibility in Arabidopsis by Enhancing Abscisic Acid (ABA) Biosynthesis. Mol. Plant Pathol. 2015, 17, 412–426. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Jin, L.P.; Xie, K.Y.; Qu, D.Y. The Potato StLTPa7 Gene Displays a Complex Ca2+-associated Pattern of Expression during the Early Stage of Potato–Ralstonia Solanacearum Interaction. Mol. Plant Pathol. 2009, 10, 15–27. [Google Scholar] [CrossRef]
- Gasser, M.; Keller, J.; Fournier, P.; Pujic, P.; Normand, P.; Boubakri, H. Identification and Evolution of nsLTPs in the Root Nodule Nitrogen Fixation Clade and Molecular Response of Frankia to AgLTP24. Sci. Rep. 2023, 13, 16020. [Google Scholar] [CrossRef]
- Chou, M.-X.; Wei, X.-Y.; Chen, D.-S.; Zhou, J.-C. Thirteen Nodule-Specific or Nodule-Enhanced Genes Encoding Products Homologous to Cysteine Cluster Proteins or Plant Lipid Transfer Proteins Are Identified in Astragalus Sinicus L. by Suppressive Subtractive Hybridization. J. Exp. Bot. 2006, 57, 2673–2685. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Chen, L.; Shi, X.; Li, Y.; Wang, J.; Chen, D.; Xie, F.; Li, Y. A Nodule-Specific Lipid Transfer Protein AsE246 Participates in Transport of Plant-Synthesized Lipids to Symbiosome Membrane and Is Essential for Nodule Organogenesis in Chinese Milk Vetch. Plant Physiol. 2014, 164, 1045–1058. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Xu, P.; Li, H.; Xu, K.; Cui, X.; Liu, Z.; Wang, X. Genetic Variation in the Glycine-Rich Protein Gene BnGRP1 Contributes to Low Phosphorus Tolerance in Brassica Napus. J. Exp. Bot. 2023, 74, 3531–3543. [Google Scholar] [CrossRef] [PubMed]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional Diversity of the Plant Glycine-Rich Proteins Superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Guo, M.; Jeong, B.; Tian, F.; Elthon, T.E.; Cerny, R.L.; Staiger, D.; Alfano, J.R. A Type III Effector ADP-Ribosylates RNA-Binding Proteins and Quells Plant Immunity. Nature 2007, 447, 284–288. [Google Scholar] [CrossRef] [PubMed]
- de Souza Cândido, E.; Pinto, M.F.S.; Pelegrini, P.B.; Lima, T.B.; Silva, O.N.; Pogue, R.; Grossi-de-Sá, M.F.; Franco, O.L. Plant Storage Proteins with Antimicrobial Activity: Novel Insights into Plant Defense Mechanisms. FASEB J. 2011, 25, 3290–3305. [Google Scholar] [CrossRef]
- Kevei, Z.; Vinardell, J.M.; Kiss, G.B.; Kondorosi, A.; Kondorosi, E. Glycine-Rich Proteins Encoded by a Nodule-Specific Gene Family Are Implicated in Different Stages of Symbiotic Nodule Development in Medicago spp. Mol. Plant Microbe Interact. 2002, 15, 922–931. [Google Scholar] [CrossRef]
- Alunni, B.; Kevei, Z.; Redondo-Nieto, M.; Kondorosi, A.; Mergaert, P.; Kondorosi, E. Genomic Organization and Evolutionary Insights on GRP and NCR Genes, Two Large Nodule-Specific Gene Families in Medicago Truncatula. Mol. Plant Microbe Interact. 2007, 20, 1138–1148. [Google Scholar] [CrossRef]
- Graham, M.A.; Silverstein, K.A.T.; Cannon, S.B.; VandenBosch, K.A. Computational Identification and Characterization of Novel Genes from Legumes. Plant Physiol. 2004, 135, 1179–1197. [Google Scholar] [CrossRef]
- Laporte, P.; Satiat-Jeunemaître, B.; Velasco, I.; Csorba, T.; de Velde, W.V.; Campalans, A.; Burgyan, J.; Arevalo-Rodriguez, M.; Crespi, M. A Novel RNA-binding Peptide Regulates the Establishment of the Medicago Truncatula–Sinorhizobium Meliloti Nitrogen-fixing Symbiosis. Plant J. 2010, 62, 24–38. [Google Scholar] [CrossRef]
- Trujillo, D.I.; Silverstein, K.A.T.; Young, N.D. Genomic Characterization of the LEED..PEEDs, a Gene Family Unique to the Medicago Lineage. G3 Genes Genomes Genet. 2014, 4, 2003–2012. [Google Scholar] [CrossRef]
- Wen, J.; Lease, K.A.; Walker, J.C. DVL, a Novel Class of Small Polypeptides: Overexpression Alters Arabidopsis Development. Plant J. 2004, 37, 668–677. [Google Scholar] [CrossRef]
- Narita, N.N.; Moore, S.; Horiguchi, G.; Kubo, M.; Demura, T.; Fukuda, H.; Goodrich, J.; Tsukaya, H. Overexpression of a Novel Small Peptide ROTUNDIFOLIA4 Decreases Cell Proliferation and Alters Leaf Shape in Arabidopsis Thaliana. Plant J. 2004, 38, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ma, Y.; Xu, Y.; Lu, P.; Yang, J.; Xie, Y.; Gan, J.; Li, L. Shade-Induced RTFL/DVL Peptides Negatively Regulate the Shade Response by Directly Interacting with BSKs in Arabidopsis. Nat. Commun. 2023, 14, 6898. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Hu, X.; Ren, M.; Ma, F.; Fu, J.; Cui, H. Stem-Cell-Expressed DEVIL-like Small Peptides Maintain Root Growth under Abiotic Stress via Abscisic Acid Signaling. Plant Physiol. 2024, 194, 2372–2386. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, H.; Kinoshita, E.; Ridge, R.W.; Kouchi, H. RNAi Knock-Down of ENOD40 s Leads to Significant Suppression of Nodule Formation in Lotus Japonicus. Plant Cell Physiol. 2006, 47, 1102–1111. [Google Scholar] [CrossRef]
- Wan, X.; Hontelez, J.; Lillo, A.; Guarnerio, C.; van de Peut, D.; Fedorova, E.; Bisseling, T.; Franssen, H. Medicago Truncatula ENOD40-1 and ENOD40-2 Are Both Involved in Nodule Initiation and Bacteroid Development. J. Exp. Bot. 2007, 58, 2033–2041. [Google Scholar] [CrossRef]
- Gultyaev, A.P.; Koster, C.; van Batenburg, D.C.; Sistermans, T.; van Belle, N.; Vijfvinkel, D.; Roussis, A. Conserved Structured Domains in Plant Non-Coding RNA Enod40, Their Evolution and Recruitment of Sequences from Transposable Elements. NAR Genom. Bioinform. 2023, 5, lqad091. [Google Scholar] [CrossRef]
- Röhrig, H.; Schmidt, J.; Miklashevichs, E.; Schell, J.; John, M. Soybean ENOD40 Encodes Two Peptides That Bind to Sucrose Synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 1915–1920. [Google Scholar] [CrossRef]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a Short Open Reading Frame-Containing mRNA, Induces Cytoplasmic Localization of a Nuclear RNA Binding Protein in Medicago Truncatula. Plant Cell 2004, 16, 1047–1059. [Google Scholar] [CrossRef]
- Ganguly, P.; Roy, D.; Das, T.; Kundu, A.; Cartieaux, F.; Ghosh, Z.; DasGupta, M. The Natural Antisense Transcript DONE40 Derived from the lncRNA ENOD40 Locus Interacts with SET Domain Protein ASHR3 During Inception of Symbiosis in Arachis Hypogaea. MPMI 2021, 34, 1057–1070. [Google Scholar] [CrossRef]
- Xu, H.; Li, Y.; Zhang, K.; Li, M.; Fu, S.; Tian, Y.; Qin, T.; Li, X.; Zhong, Y.; Liao, H. miR169c-NFYA-C-ENOD40 Modulates Nitrogen Inhibitory Effects in Soybean Nodulation. New Phytol. 2021, 229, 3377–3392. [Google Scholar] [CrossRef]
- Li, X.; Zhou, H.; Cheng, L.; Ma, N.; Cui, B.; Wang, W.; Zhong, Y.; Liao, H. Shoot-to-Root Translocated GmNN1/FT2a Triggers Nodulation and Regulates Soybean Nitrogen Nutrition. PLoS Biol. 2022, 20, e3001739. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, C.; Charon, C.; Boller, T.; Crespi, M.; Kondorosi, Á. Medicago Truncatula Plants Overexpressing the Early Nodulin Gene Enod40 Exhibit Accelerated Mycorrhizal Colonization and Enhanced Formation of Arbuscules. Proc. Natl. Acad. Sci. USA 2001, 98, 15366–15371. [Google Scholar] [CrossRef]
- Grundy, E.B.; Gresshoff, P.M.; Su, H.; Ferguson, B.J. Legumes Regulate Symbiosis with Rhizobia via Their Innate Immune System. Int. J. Mol. Sci. 2023, 24, 2800. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E.D. Plant Signalling in Symbiosis and Immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An Integrated Transcriptome Atlas of the Crop Model Glycine Max, and Its Use in Comparative Analyses in Plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Tzipilevich, E.; Russ, D.; Dangl, J.L.; Benfey, P.N. Plant Immune System Activation Is Necessary for Efficient Root Colonization by Auxin-Secreting Beneficial Bacteria. Cell Host Microbe 2021, 29, 1507–1520.e4. [Google Scholar] [CrossRef] [PubMed]
- Gully, D.; Czernic, P.; Cruveiller, S.; Mahé, F.; Longin, C.; Vallenet, D.; François, P.; Nidelet, S.; Rialle, S.; Giraud, E.; et al. Transcriptome Profiles of Nod Factor-Independent Symbiosis in the Tropical Legume Aeschynomene Evenia. Sci. Rep. 2018, 8, 10934. [Google Scholar] [CrossRef]
- Ding, S.; Lv, J.; Hu, Z.; Wang, J.; Wang, P.; Yu, J.; Foyer, C.H.; Shi, K. Phytosulfokine Peptide Optimizes Plant Growth and Defense via Glutamine Synthetase GS2 Phosphorylation in Tomato. EMBO J. 2023, 42, e111858. [Google Scholar] [CrossRef]
- A Fungal Pathogen Manipulates Phytocytokine Signaling for Plant Infection|bioRxiv. Available online: https://www.biorxiv.org/content/10.1101/2023.07.04.547642v1 (accessed on 12 November 2024).
- Dhar, N.; Caruana, J.; Erdem, I.; Raina, R. An Arabidopsis DISEASE RELATED NONSPECIFIC LIPID TRANSFER PROTEIN 1 Is Required for Resistance against Various Phytopathogens and Tolerance to Salt Stress. Gene 2020, 753, 144802. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, C.; Shan, R.; Li, X.; Tseke Inkabanga, A.; Li, L.; Jiang, H.; Chai, Y. Genome-Wide Identification and Expression Analysis of nsLTP Gene Family in Rapeseed (Brassica Napus) Reveals Their Critical Roles in Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 8372. [Google Scholar] [CrossRef] [PubMed]
- Bobay, B.G.; DiGennaro, P.; Scholl, E.; Imin, N.; Djordjevic, M.A.; Mck Bird, D. Solution NMR Studies of the Plant Peptide Hormone CEP Inform Function. FEBS Lett. 2013, 587, 3979–3985. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Furumizu, C.; Zhang, B.; Sawa, S. Database Mining of Plant Peptide Homologues. Plant Biotechnol. 2021, 38, 137–143. [Google Scholar] [CrossRef]
- Thoms, D.; Liang, Y.; Haney, C.H. Maintaining Symbiotic Homeostasis: How Do Plants Engage With Beneficial Microorganisms While at the Same Time Restricting Pathogens? MPMI 2021, 34, 462–469. [Google Scholar] [CrossRef]
- Farkas, A.; Maróti, G.; Dürgő, H.; Györgypál, Z.; Lima, R.M.; Medzihradszky, K.F.; Kereszt, A.; Mergaert, P.; Kondorosi, É. Medicago Truncatula Symbiotic Peptide NCR247 Contributes to Bacteroid Differentiation through Multiple Mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 5183–5188. [Google Scholar] [CrossRef] [PubMed]
- Dodueva, I.; Lebedeva, M.; Lutova, L. Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones. Plants 2021, 10, 2243. [Google Scholar] [CrossRef]
- Mondino, S.; Schmidt, S.; Buchrieser, C. Molecular Mimicry: A Paradigm of Host-Microbe Coevolution Illustrated by Legionella. mBio 2020, 11, e01201-20. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yang, R.; Chang, C.; Park, Y.; Tucker, M.L. The Root-Knot Nematode Meloidogyne Incognita Produces a Functional Mimic of the Arabidopsis INFLORESCENCE DEFICIENT IN ABSCISSION Signaling Peptide. J. Exp. Bot. 2018, 69, 3009–3021. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Liu, X. Peptide Effectors in Phytonematode Parasitism and Beyond. Annu. Rev. Phytopathol. 2022, 60, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Thynne, E.; Saur, I.M.L.; Simbaqueba, J.; Ogilvie, H.A.; Gonzalez-Cendales, Y.; Mead, O.; Taranto, A.; Catanzariti, A.-M.; McDonald, M.C.; Schwessinger, B.; et al. Fungal Phytopathogens Encode Functional Homologues of Plant Rapid Alkalinization Factor (RALF) Peptides. Mol Plant Pathol. 2017, 18, 811–824. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Joe, A.; Zhang, W.; Feng, W.; Stewart, V.; Schwessinger, B.; Dinneny, J.R.; Ronald, P.C. A Microbially Derived Tyrosine-sulfated Peptide Mimics a Plant Peptide Hormone. New Phytol. 2017, 215, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Chen, X.; Kemppainen, M.; Pardo, A.G.; Veneault-Fourrey, C.; Kohler, A.; Martin, F.M. The Small Secreted Effector Protein MiSSP7.6 of Laccaria Bicolor Is Required for the Establishment of Ectomycorrhizal Symbiosis. Environ. Microbiol. 2020, 22, 1435–1446. [Google Scholar] [CrossRef]
- Kamel, L.; Tang, N.; Malbreil, M.; San Clemente, H.; Le Marquer, M.; Roux, C.; Frei dit Frey, N. The Comparison of Expressed Candidate Secreted Proteins from Two Arbuscular Mycorrhizal Fungi Unravels Common and Specific Molecular Tools to Invade Different Host Plants. Front. Plant Sci. 2017, 8, 124. [Google Scholar] [CrossRef]
- Foo, E. Something Old, Something New: Auxin and Strigolactone Interact in the Ancient Mycorrhizal Symbiosis. Plant Signal. Behav. 2013, 8, e23656. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.M.; Harrison, M.J. Phytohormones, miRNAs, and Peptide Signals Integrate Plant Phosphorus Status with Arbuscular Mycorrhizal Symbiosis. Curr. Opin. Plant Biol. 2019, 50, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Greifenhagen, A.; Ruwe, H.; Zimmer, V.; Messerschmidt, J.; Bhukya, D.P.N.; Kenea, H.D.; Schaller, A.; Spallek, T. The Peptide Hormone PjCLE1 Stimulates Haustorium Formation in the Parasitic Plant Phtheirospermum Japonicum. Proc. Natl. Acad. Sci. USA 2024, 121, e2414582121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Ye, Q.; Chen, H.; Dong, J.; Wang, T. Multigene Editing Reveals That MtCEP1/2/12 Redundantly Control Lateral Root and Nodule Number in Medicago Truncatula. J. Exp. Bot. 2021, 72, 3661–3676. [Google Scholar] [CrossRef]
- Jain, D.; Jones, L.; Roy, S. Gene Editing to Improve Legume-Rhizobia Symbiosis in a Changing Climate. Curr. Opin. Plant Biol. 2023, 71, 102324. [Google Scholar] [CrossRef]

| Peptide Family | Nodules | Mycorrhiza | PGPB | Produced by Pathogens | Root Growth | Immunity | References |
|---|---|---|---|---|---|---|---|
| CLE | + | + | + | + | [28,36,48,49] | ||
| CEP | + | + | + | + | [69,70,72,177] | ||
| RALF | + | + | + | + | + | [124,125,129,130,135,136,137] | |
| PSK | + | + (PSKR1) | + | + | + | [82,83,84,88,178] | |
| nsLTP | + | + | + | [139,142,175] | |||
| GRP | + | + | + | [146,148,150] | |||
| RGF/GLV | + | + | + | [77,79,81] | |||
| DVL | + | + | [130,158] | ||||
| DEF | + | + | [93,94] | ||||
| NCR | + | [7,9,10] | |||||
| SNARP/LP | + | [153] | |||||
| ENOD40 | + | [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamaeva, A.; Makeeva, A.; Ganaeva, D. The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions. Plants 2025, 14, 378. https://doi.org/10.3390/plants14030378
Mamaeva A, Makeeva A, Ganaeva D. The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions. Plants. 2025; 14(3):378. https://doi.org/10.3390/plants14030378
Chicago/Turabian StyleMamaeva, Anna, Arina Makeeva, and Daria Ganaeva. 2025. "The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions" Plants 14, no. 3: 378. https://doi.org/10.3390/plants14030378
APA StyleMamaeva, A., Makeeva, A., & Ganaeva, D. (2025). The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions. Plants, 14(3), 378. https://doi.org/10.3390/plants14030378






