Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms
Abstract
1. Introduction
2. Results
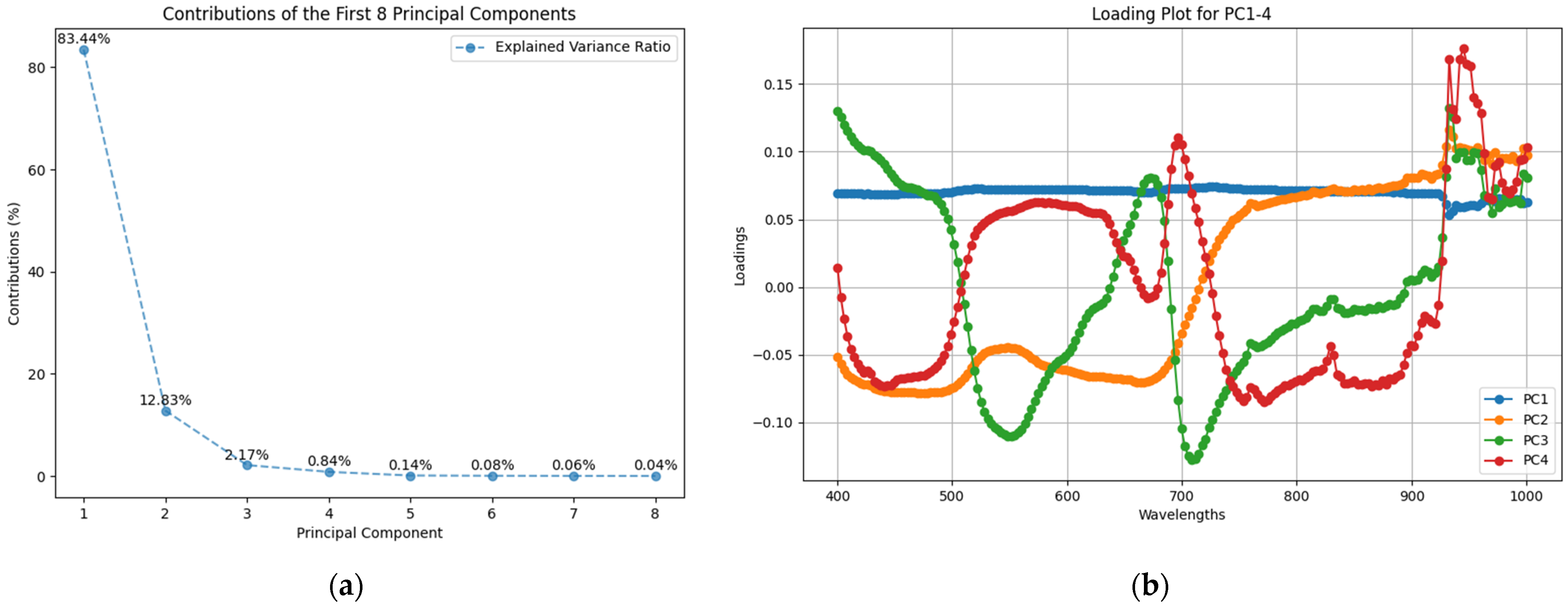
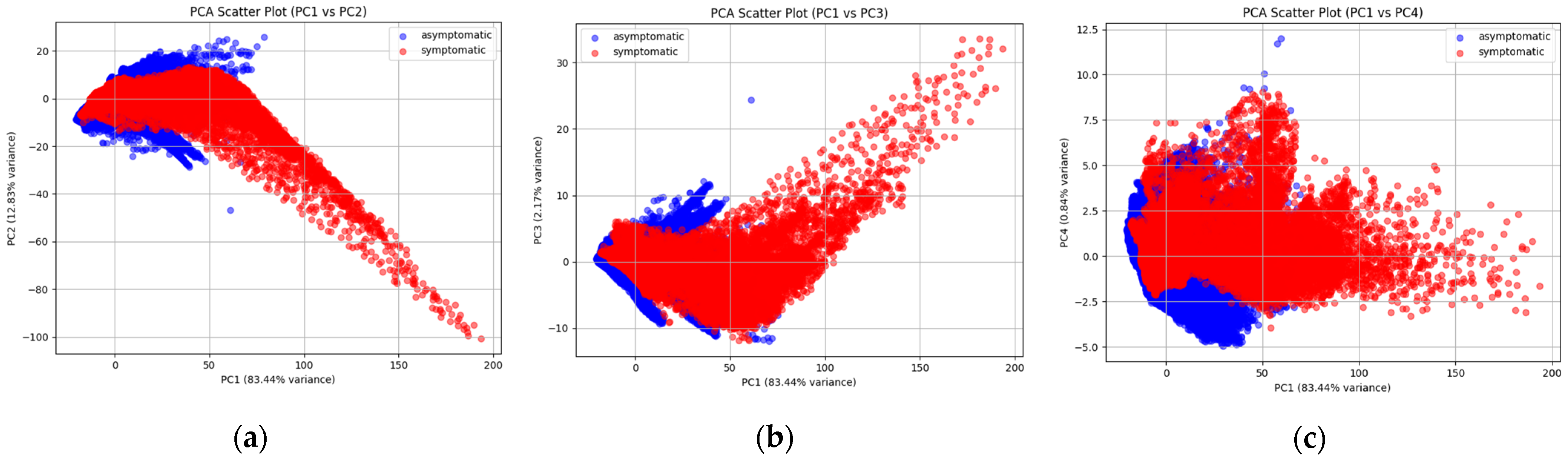
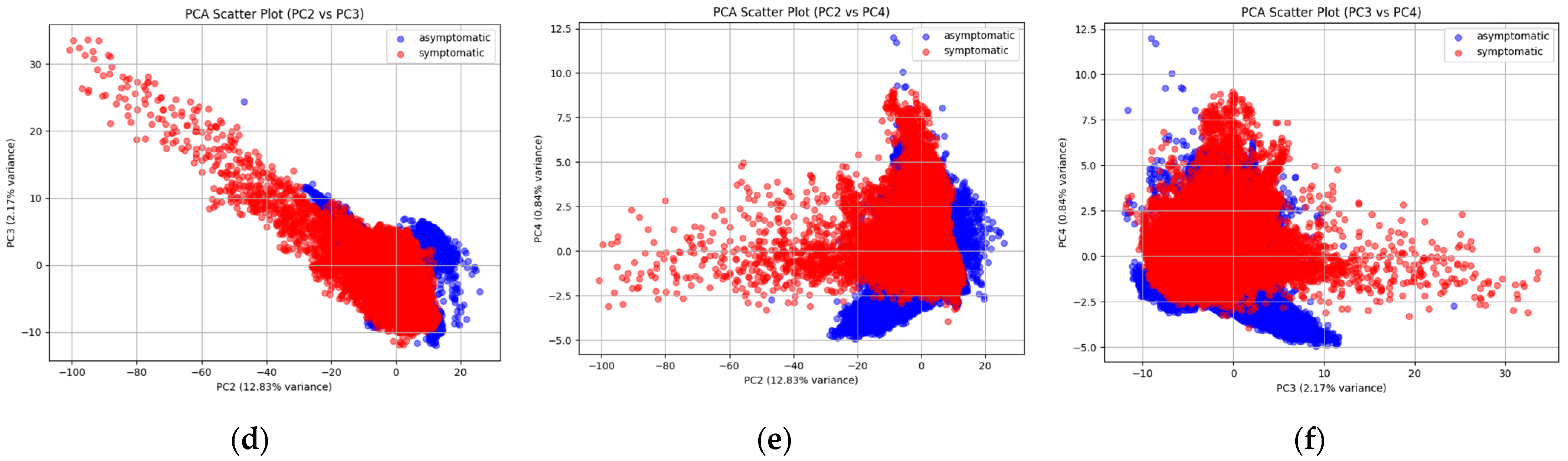
2.1. PCA for the Explanation of the Variance in the Data
2.2. Hyperspectral Data Wavelength Optimization
2.3. Reliability of Expert System
3. Discussion
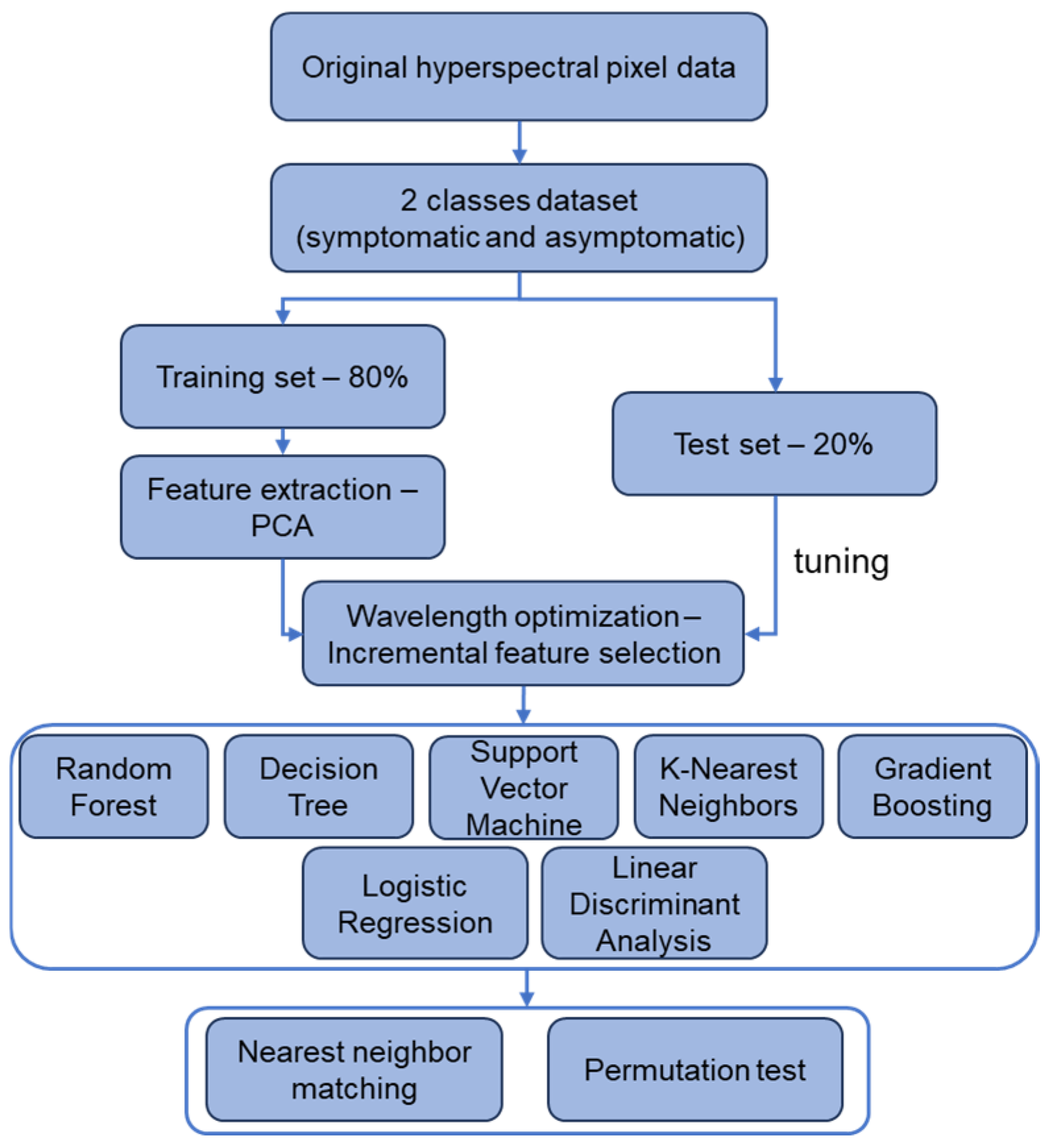
4. Materials and Methods
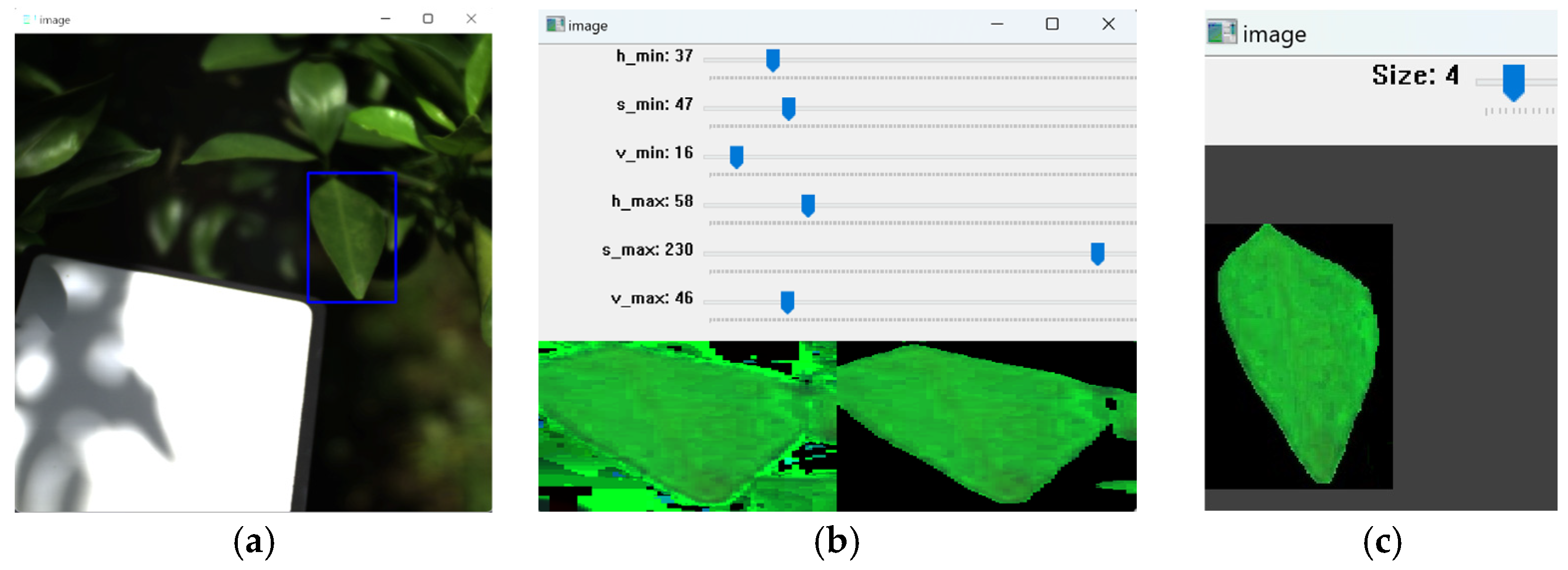
4.1. Hyperspectral Image Data Collection
4.2. Hyperspectral Data Preprocessing
4.3. Hyperspectral Data Analysis and Modeling
4.3.1. Feature Extraction—PCA
4.3.2. Wavelength Optimization—Incremental Feature Selection
4.3.3. Machine Learning Models
4.4. Evaluation of Models for Leaf Image Separation
4.4.1. F1 Score
4.4.2. Nearest Neighbor Matching
4.4.3. Permutation Test
5. Conclusions
- -
- Our RF, decision tree, and KNN models are as reliable as PCR in identifying HLB.
- -
- Nonlinear models outperform linear models for HLB spectral data.
- -
- Using PCA for nonlinear models is effective for HLB feature extraction.
- -
- Decision tree model provides high accuracy with faster prediction, suitable for real-time applications.
- -
- KNN model shows promising potential for multispectral imaging applications.
- -
- The red-edge and near-infrared regions may be critical for HLB detection.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tipu, M.M.H.; Masud, M.M.; Jahan, R.; Baroi, A.; Hoque, A.K.M.A. Identification of citrus greening based on visual symptoms: A grower’s diagnostic toolkit. Heliyon 2021, 7, e08387. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Lan, Y.; Hong, T.; Chen, J. Citrus greening detection using visible spectrum imaging and C-SVC. Compt. Electron. Agric. 2016, 130, 177–183. [Google Scholar] [CrossRef]
- Cap, H.Q.; Suwa, K.; Fujita, E.; Kagiwada, S.; Uga, H.; Iyatomi, H. A deep learning approach for on-site plant leaf detection. In Proceedings of the 2018 IEEE 14th International Colloquium on Signal Processing & Its Applications, Penang, Malaysia, 9–10 March 2018; pp. 18–122. [Google Scholar]
- Yang, D.; Wang, F.; Hu, Y.; Lan, Y.; Deng, X. Citrus huanglongbing detection based on multi-modal feature fusion learning. Front. Plant Sci. 2021, 12, 809506. [Google Scholar] [CrossRef] [PubMed]
- Gottwald, T.R. Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef]
- Kalim, H.; Chug, A.; Singh, A.P. Citrus leaf disease detection using hybrid CNN-RF model. In Proceedings of the 4th International Conference on Artificial Intelligence and Speech Technology (AIST), Delhi, India, 9–10 December 2022; pp. 1–4. [Google Scholar]
- Elaraby, A.; Hamdy, W.; Alanazi, S. Classification of citrus diseases using optimization deep learning approach. Comput. Intell. Neurosci. 2022, 2022, 9153207. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Flores, W.; Garza-Saldaña, J.J.; Varela-Fuentes, S.E. A huanglongbing detection method for orange trees based on deep neural networks and transfer learning. IEEE Access 2022, 10, 116686–116696. [Google Scholar] [CrossRef]
- Liu, X.; Min, W.; Mei, S.; Wang, L.; Jiang, S. Plant disease recognition: A large-scale benchmark dataset and a visual region and loss reweighting approach. IEEE Transact. Image Process. 2021, 30, 2003–2015. [Google Scholar] [CrossRef]
- Xia, F.; Xie, X.; Wang, Z.; Jin, S.; Yan, K.; Ji, Z. A novel computational framework for precision diagnosis and subtype discovery of plant with lesion. Front. Plant Sci. 2022, 12, 789630. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Dongmei, G.; Zhang, Y.; Deng, L.; Xie, R.; Lv, Q.; Yi, S.; Zheng, Y.; Ma, Y.; He, S. Detection of Huanglongbing (citrus greening) based on hyperspectral image analysis and PCR. Front. Agric. Sci. Eng. 2019, 6, 172–180. [Google Scholar] [CrossRef]
- Lan, Y.; Huang, Z.; Deng, X.; Zhu, Z.; Huang, H.; Zheng, Z.; Lian, B.; Zeng, G.; Tong, T. Comparison of machine learning methods for citrus greening detection on UAV multispectral images. Compt. Electron. Agric. 2020, 171, 105234. [Google Scholar] [CrossRef]
- Deng, X.; Huang, Z.; Zheng, Z.; Lan, Y.; Dai, F. Field detection and classification of citrus Huanglongbing based on hyperspectral reflectance. Compt. Electron. Agric. 2019, 167, 105006. [Google Scholar] [CrossRef]
- Podlesnykh, I.; Kovalev, M.; Platonov, P. Towards the future of ubiquitous hyperspectral imaging: Innovations in sensor configurations and cost reduction for widespread applicability. Technologies 2024, 12, 221. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Maja, J.M.; Ehsani, R. Visible-near infrared spectroscopy for detection of Huanglongbing (HLB) Using a VIS-NIR Spectroscopy Technique. Comput. Electron. Agric. 2011, 77, 127–134. [Google Scholar] [CrossRef]
- Sankaran, S.; Ehsani, R. Visible-near infrared spectroscopy based citrus greening detection: Evaluation of spectral feature extraction techniques. Crop Prot. 2011, 30, 1508–1513. [Google Scholar] [CrossRef]
- Sankaran, S.; Maja, J.; Buchanon, S.; Ehsani, R. Huanglongbing (Citrus Greening) Detection Using Visible, Near Infrared and Thermal Imaging Techniques. Sensors 2013, 13, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lee, W.S.; Li, M.; Ehsani, R.; Mishra, A.R.; Yang, C.; Mangan, R.L. Spectral difference analysis and airborne imaging classification for citrus greening infected trees. Comput. Electron. Agric. 2012, 83, 32–46. [Google Scholar] [CrossRef]
- Kumar, A.; Lee, W.S.; Ehsani, R.J.; Albrigo, L.G.; Yang, C.; Mangane, R.L. Citrus greening disease detection using aerial hyperspectral and multispectral imaging techniques. J. Appl. Remote Sens. 2012, 6, 063542. [Google Scholar]
- Weng, H.; Lu, J.; Cen, H.; He, M.; Zeng, Y.; Hua, S.; Li, H.; Meng, Y.; Fang, H.; He, Y. Hyperspectral reflectance imaging combined with carbohydrate metabolism analysis for diagnosis of citrus Huanglongbing in different seasons and cultivars. Sens. Actuators B Chem. 2018, 275, 50–60. [Google Scholar] [CrossRef]
- Deng, X.; Zhu, Z.; Yang, J.; Zheng, Z.; Huang, Z.; Yin, X.; Wei, S.; Lan, Y. Detection of Citrus Huanglongbing Based on Multi-Input Neural Network Model of UAV HRS. Remote Sens. 2020, 12, 2678. [Google Scholar] [CrossRef]
- Menezes, J.; Dharmalingam, R.; Shivakumara, P. HLB Disease Detection in Omani Lime Trees Using Hyperspectral Imaging Based Techniques. In Recent Trends in Image Processing and Pattern Recognition; Springer Nature: Cham, Switzerland, 2024; pp. 67–81. [Google Scholar]
- Yan, K.; Song, X.; Yang, J.; Xiao, J.; Xu, X.; Guo, J.; Zhu, H.; Lan, Y.; Zhang, Y. Citrus huanglongbing detection: A hyperspectral data-driven model integrating feature band selection with machine learning algorithms. Crop Prot. 2025, 188, 107008. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Ustin, S.L.; Jacquemoud, S. How the optical properties of leaves modify the absorption and scattering of energy and enhance leaf functionality. In Remote Sensing of Plant Biodiversity; Cavender-Bares, J., Gamon, J.A., Townsend, P.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 349–384. [Google Scholar]
- Peñuelas, J.; Inoue, Y. Reflectance indices indicative of changes in water and pigment contents of peanut and wheat leaves. Photosynthetica 1999, 36, 355–360. [Google Scholar] [CrossRef]
- Zheng, J.; Rakovski, C. On the Application of Principal Component Analysis to Classification Problems. Data Sci. J. 2021, 20, 26. [Google Scholar] [CrossRef]
- Silva, R.; Melo-Pinto, P. t-SNE: A study on reducing the dimensionality of hyperspectral data for the regression problem of estimating oenological parameters. Artif. Intell. Agric. 2023, 7, 58–68. [Google Scholar] [CrossRef]
- Kang, Z.; Fan, R.; Zhan, C.; Wu, Y.; Lin, Y.; Li, K.; Qing, R.; Xu, L. The Rapid Non-Destructive Differentiation of Different Varieties of Rice by Fluorescence Hyperspectral Technology Combined with Machine Learning. Molecules 2024, 29, 682. [Google Scholar] [CrossRef]
- Kobayash, O.; Dong, R.; Shiraiwa, A. Eye-tracking data analysis for improving diagnosis performance in citrus greening disease. IEICE Tech. Rep. 2024, 124, 35–38. [Google Scholar]
- Dong, R.; Shiraiwa, A.; Pawasut, A.; Sreechun, K.; Hayashi, T. Diagnosis of Citrus Greening Using Artificial Intelligence: A Faster Region-Based Convolutional Neural Network Approach with Convolution Block Attention Module-Integrated VGGNet and ResNet Models. Plants 2024, 13, 1631. [Google Scholar] [CrossRef]
- Rosenblatt, M.; Tejavibulya, L.; Jiang, R.; Noble, S.; Scheinost, D. Data leakage inflates prediction performance in connectome-based machine learning models. Nat. Comm. 2024, 15, 1829. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal component analysis. In International Encyclopedia of Statistical Science; Lovric, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1094–1096. [Google Scholar]
- Liu, H.; Setiono, R. Incremental feature selection. Appl. Intell. 1998, 9, 217–230. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 1984. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Jerome, H. Friedman. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Kleinbaum, D.G. Logistic Regression: A Self-Learning Text; Springer: Berlin/Heidelberg, Germany, 1994. [Google Scholar]
- Liu, Z.P. Linear Discriminant Analysis. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 1132–1133. [Google Scholar] [CrossRef]
- Powers, D. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness & correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Clark, P.J.; Evans, F.C. Distance to Nearest Neighbor as a Measure of Spatial Relationships in Populations. Ecology 1954, 35, 445–453. [Google Scholar] [CrossRef]
- Ojala, M.; Garriga, G.C. Permutation tests for studying classifier performance. In Proceedings of the 2009 Ninth IEEE International Conference on Data Mining, Miami, FL, USA, 6–9 December 2009; pp. 908–913. [Google Scholar]




| Publication Year | Equipment Wavelengths (nm) | Feature Wavelengths (nm) | Study Type |
|---|---|---|---|
| 2011–2013 [16,17,18] | 350–2500 | 537, 612, 638, 662, 688, 713, 763, 813, 998, 1066, 1120, 1148, 1296, 1445, 1472, 1546, 1597, 1622, 1746, 1898, 2121, 2172, 2348, 2471, 2493 | field |
| 2012 [19] | 457–921 | 650–850 | field and lab |
| 2012 [20] | 457–921 | 410–432, 440–509, 634–686, 734–927, 932, 951, 975, 980 | field |
| 2018 [21] | 379–1023 | 493, 515, 665, 716, 739 | lab |
| 2019 [14] | 400–1000 | 544, 718, 753, 760, 764, 930, 938, 943, 951, 969, 985, 998, 999 | field |
| 2020 [22] | 450–950, 325–1075 | 468, 504, 512, 516, 528, 536, 632, 680, 688, 852 | field |
| 2024 [23] | 400–1000 | 560, 678, 726, 750 | lab |
| 2025 [24] | 325–1075 | 375–425, 650–750, 890–925 | lab |
| PC1 | PC2 | PC3 | PC4 | ||||
|---|---|---|---|---|---|---|---|
| Wavelength | Loading | Wavelength | Loading | Wavelength | Loading | Wavelength | Loading |
| 727 | 0.0741 | 933 | 0.1165 | 933 | 0.1324 | 945 | 0.1764 |
| 724 | 0.0741 | 936 | 0.1111 | 400 | 0.1303 | 942 | 0.1686 |
| 730 | 0.0740 | 930 | 0.1040 | 709 | −0.1276 | 933 | 0.1683 |
| 721 | 0.0739 | 957 | 0.1032 | 712 | −0.1271 | 948 | 0.1648 |
| 733 | 0.0739 | 942 | 0.1031 | 936 | 0.1260 | 951 | 0.1636 |
| 718 | 0.0738 | 939 | 0.1025 | 403 | 0.1254 | 954 | 0.1400 |
| 736 | 0.0737 | 945 | 0.1025 | 706 | −0.1247 | 957 | 0.1359 |
| 715 | 0.0735 | 997 | 0.1025 | 715 | −0.1232 | 936 | 0.1312 |
| Classification Model | Full Wavelengths (204 Wavelengths) | Feature Extraction (16 Wavelengths) | Wavelength Optimization (No. of Wavelengths) | |
|---|---|---|---|---|
| Nonlinear | RF | 99.5% | 99.6% | 99.8% (9) |
| Decision tree | 99.1% | 99.2% | 99.3% (6) | |
| KNN | 98.8% | 96.3% | 97.9% (4) | |
| Gradient boosting | 97.7% | 95.6% | 96.2% (12) | |
| SVM | 98.4% | 89.2% | 89.2% (16) | |
| Linear | LDA | 89.3% | 38.8% | 38.8% (15) |
| Logistic regression | 91.8% | 38.3% | 38.4% (14) | |
| Classification Model | Extracted Wavelengths (nm) | Discrepancies |
|---|---|---|
| Deng et al. [14] | 544, 718, 753, 760, 764, 930, 938, 943, 951, 969, 985, 998, 999 | |
| RF | 727, 930, 933, 936, 939, 942, 945, 957, 997 | 2.78 |
| Decision tree | 727, 930, 933, 936, 939, 942, 957 | 3.14 |
| KNN | 727, 930, 933, 936 | 3.5 |
| Gradient boosting | 721, 724, 727, 730, 733, 930, 933, 936, 939, 942, 945, 957 | 5.0 |
| Classification Model | F1-Score | Wavelengths Used | Test Time (ms) |
|---|---|---|---|
| RF | 99.8% | 9 | 869 |
| Decision tree | 99.3% | 7 | 7 |
| KNN | 97.9% | 4 | 899 |
| Device | Specification | Value |
|---|---|---|
| Specim IQ | Resolution | 512 × 512 pix |
| Wavelength range (204) | 397–1004 nm | |
| Dimension | 207 × 91 × 74 mm | |
| Pixel size | 17.58 μm × 17.58 μm | |
| Calibration whiteboard | Reflectivity | 100% |
| Size | 10 × 10 cm | |
| Neutral density filter | Average Transmission | 25% |
| PC1 | PC2 | Wavelength Selection | Counts |
|---|---|---|---|
| A1 | B1 | A1B1, A1B1B2, …, A1B1B2B3… Bn | n |
| A2 | B2 | A1A2B1, A1A2B1B2, …, A1A2B1B2B3… Bn | n |
| A3 | B3 | A1A2A3B1, A1A2A3B1B2, …, A1A2A3B1B2B3… Bn | n |
| …… | …… | n | |
| An | Bn | A1A2A3… AnB1, A1A2A3… AnB1B2, …, A1A2A3… AnB1B2B3… Bn | n |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, R.; Shiraiwa, A.; Ichinose, K.; Pawasut, A.; Sreechun, K.; Mensin, S.; Hayashi, T. Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms. Plants 2025, 14, 451. https://doi.org/10.3390/plants14030451
Dong R, Shiraiwa A, Ichinose K, Pawasut A, Sreechun K, Mensin S, Hayashi T. Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms. Plants. 2025; 14(3):451. https://doi.org/10.3390/plants14030451
Chicago/Turabian StyleDong, Ruihao, Aya Shiraiwa, Katsuya Ichinose, Achara Pawasut, Kesaraporn Sreechun, Sumalee Mensin, and Takefumi Hayashi. 2025. "Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms" Plants 14, no. 3: 451. https://doi.org/10.3390/plants14030451
APA StyleDong, R., Shiraiwa, A., Ichinose, K., Pawasut, A., Sreechun, K., Mensin, S., & Hayashi, T. (2025). Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms. Plants, 14(3), 451. https://doi.org/10.3390/plants14030451






