The WRKY28-BRC1 Transcription Factor Module Controls Shoot Branching in Brassica napus
Abstract
:1. Introduction
2. Results
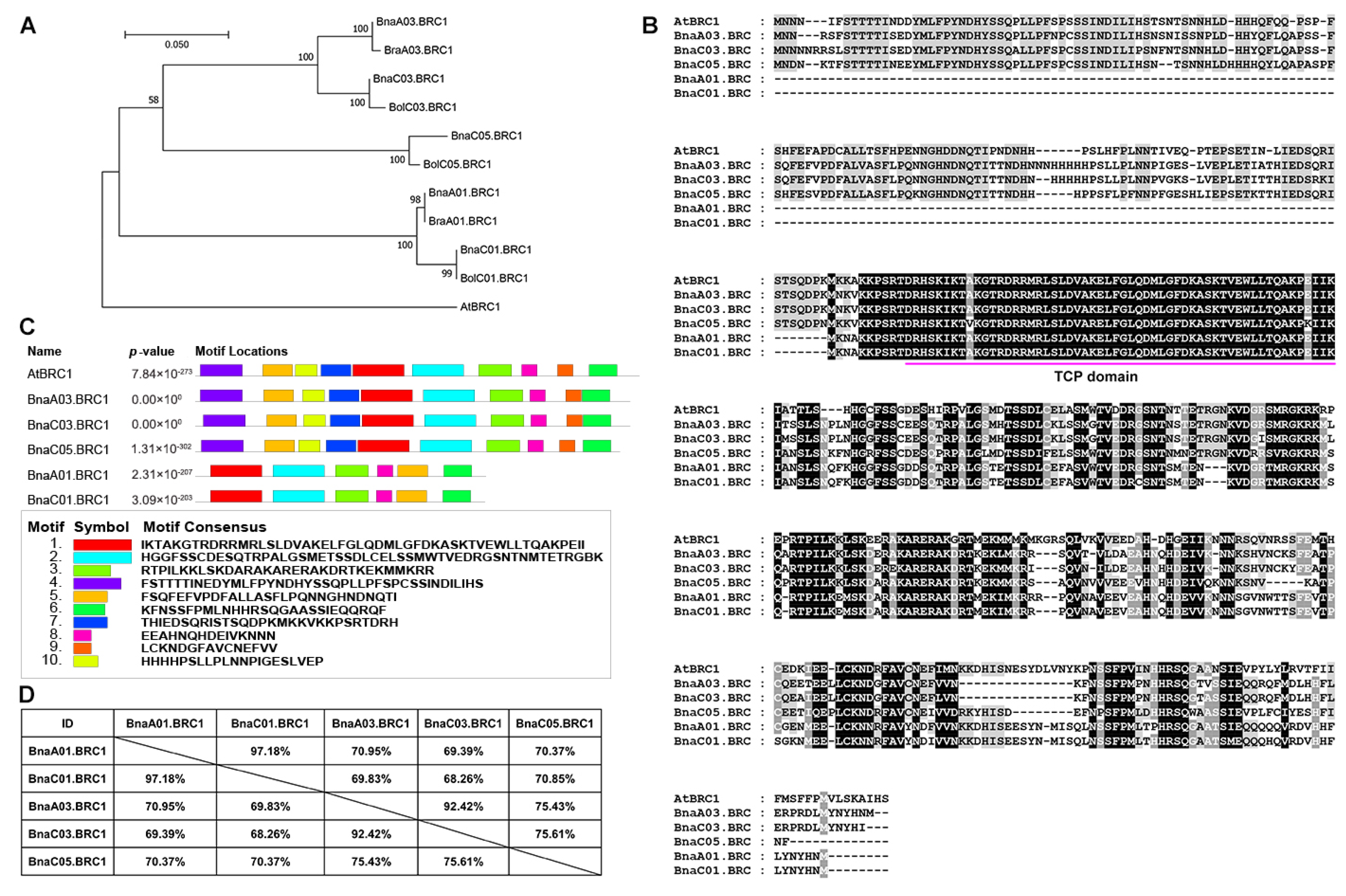
2.1. Five BRC1 Copies Are Identified in the Rapeseed Genome
2.2. Multiple Copies of BRC1 May Function Redundantly in Shoot Branching in Rapeseed
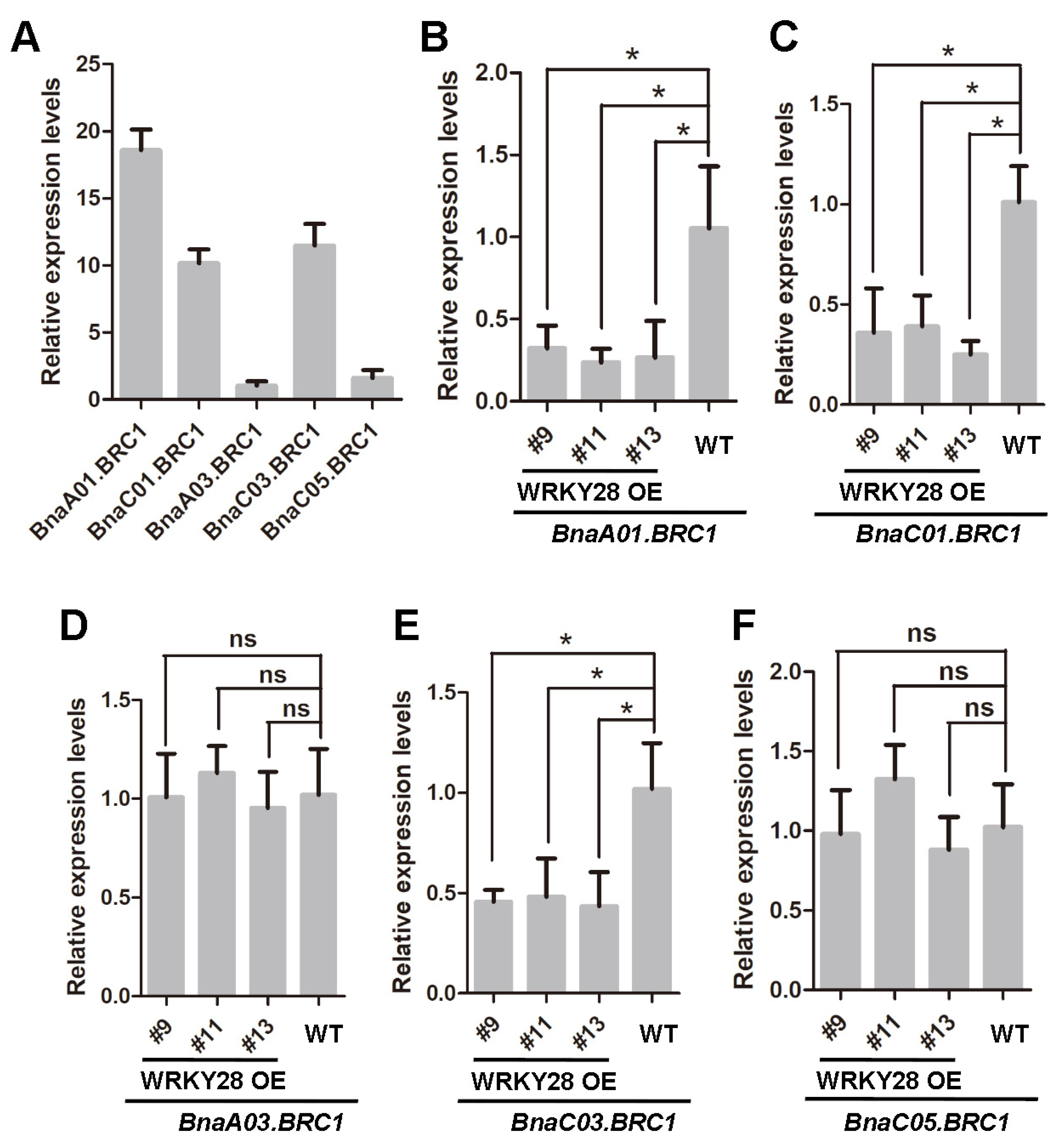
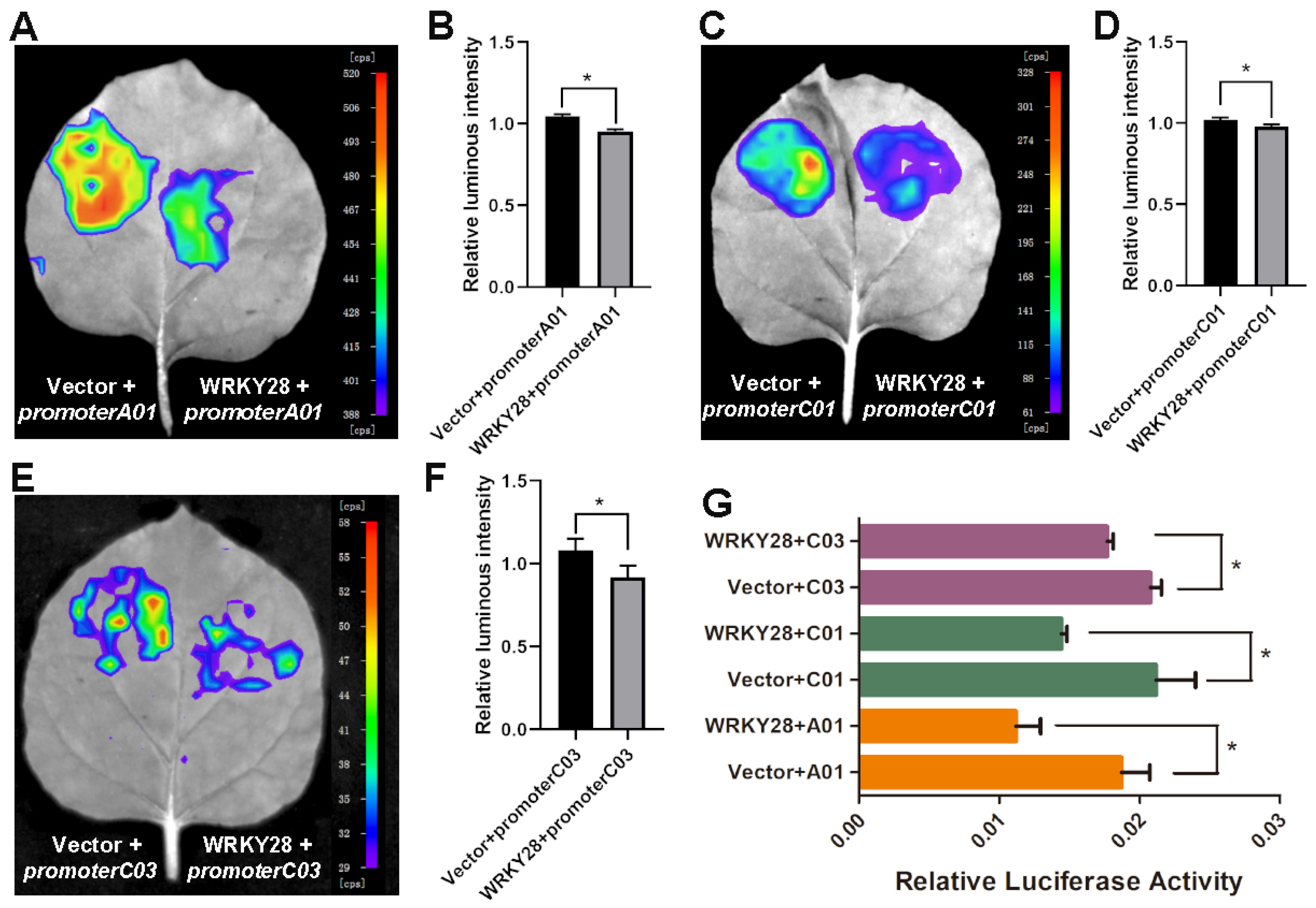
2.3. BnaA03.WRKY28 Targets BnaA01.BRC1, BnaC01.BRC1, and BnaC03.BRC1 and Negatively Regulates Their Expression
2.4. The brc1 Mutants Exhibit Excessive Axillary Branching Formation
2.5. BRC1 May Regulate Axillary Bud Dormancy and Outgrowth Through Hormone Signaling in Rapeseed
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Generation of Binary Constructs and Knockout Lines
4.4. Isolation of RNA and RT-qPCR
4.5. Electrophoretic Mobility Shift Assay (EMSA)
4.6. Dual-LUC Transient Transcriptional Activity Assay
4.7. Histological Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WT | Wild-type |
| Bn/Bna | Brassica napus |
| TCP | Teosinte branched1/Cycloidea/Proliferating cell factor |
| At | Arabidopsis thaliana |
| TF | Transcription factor |
| EMSA | Electrophoretic Mobility Shift Assay |
| LUC | Luciferase |
| AM | Axillary meristem |
| CDS | Coding sequence |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| RT-qPCR | Real-time quantitative PCR |
| IAA | Auxin |
| SL | Strigolactone |
| ABA | Abscisic acid |
| NCED | 9-cis epoxycarotenoid dioxygenase |
| CYP | Cytochrome P450 |
| LAS | LATERAL SUPPRESSOR |
| RAX | REGULATOR OF AXILLARY MERISTEMS |
| CUC | CUP-SHAPED COTYLEDON |
| BRC1 | BRANCHED 1 |
| PIN1 | PIN-FORMED1 |
| AXR1 | AUXIN RESISTENT 1 |
| MAX | MORE AXILLARY GROWTH |
| FHY3 | FAR-RED ELONGATED HYPOCOTYL3 |
References
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xu, K.; Li, J.; Li, F.; Qiao, J.; Li, H.; Gao, G.; Yan, G.; Wu, X.J. Evaluation of yield and agronomic traits and their genetic variation in 488 global collections of Brassica napus L. Genet. Resour. Crop Evol. 2014, 61, 979–999. [Google Scholar] [CrossRef]
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Axillary meristem initiation-a way to branch out. Curr. Opin. Plant Biol. 2018, 41, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Sato, S.; Mori, H. Control of outgrowth and dormancy in axillary buds. Plant Physiol. 2001, 127, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An update on the signals controlling shoot branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, W.; Yamauchi, T.; Tsuda, K. Genetic basis controlling rice plant architecture and its modifcation for breeding. Breed. Sci. 2023, 73, 3–45. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes. Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Mybgenes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.C.J.; de Vries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundaryand shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Kotov, A.A.; Kotova, L.M.; Romanov, G.A. Signaling network regulating plant branching: Recent advances and new challenges. Plant Sci. 2021, 307, 110880. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Booker, J.; Chatfield, S.; Leyser, O. Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 2003, 15, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.; Jiao, Y. The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067. [Google Scholar] [CrossRef]
- Chatfield, S.P.; Stirnberg, P.; Forde, B.G.; Leyser, O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000, 24, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Liang, Y.; Seale, M.; Ward, S.; Müller, D.; Leyser, O. Strigolactone regulates shoot development through a core signalling pathway. Biol. Open 2016, 5, 1806–1820. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.Q.; Emery, R.J.N. Is ABA the earliest upstream inhibitor of apical dominance? J. Exp. Bot. 2017, 68, 881–884. [Google Scholar] [CrossRef]
- Danisman, S.; van Dijk, A.D.J.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G.H. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef]
- Kim, S.H.; Son, G.H.; Bhattacharjee, S.; Kim, H.J.; Nam, J.C.; Nguyen, P.D.T.; Hong, J.C.; Gassmann, W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014, 78, 978–989. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Gustus, C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; Germain, A.D.S.; Pillot, J.-P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, J.; Wang, X.; Han, X.; Wei, B.; Wang, J.; Li, B.; Yu, H.; Huang, Q.; Gu, H.; et al. The WRKY Transcription Factor WRKY71/EXB1 Controls Shoot Branching by Transcriptionally Regulating RAX Genes in Arabidopsis. Plant Cell 2015, 27, 3112–3127. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cai, H.; Su, Z.; Wang, L.; Huang, X.; Zhang, M.; Chen, P.; Dai, X.; Zhao, H.; Palanivelu, R.; et al. KLU suppresses megasporocyte cell fate through SWR1-mediated activation of expression in. Proc. Natl. Acad. Sci. USA 2018, 115, E526–E535. [Google Scholar] [CrossRef]
- Tian, T.; Ma, L.; Liu, Y.; Xu, D.; Chen, Q.; Li, G. Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 Integrates Age and Light Signals to Negatively Regulate Leaf Senescence. Plant Cell 2020, 32, 1574–1588. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, F.; Wang, Z.; Zhuo, C.; Hu, K.; Li, X.; Tu, J. Transcription factor WRKY28 curbs WRKY33-mediated resistance to Sclerotinia sclerotiorum in Brassica napus. Plant Physiol. 2022, 190, 2757–2774. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Peng, F.; Tang, Q.; Liu, Y.; Xia, H.; Yao, X.; Lu, S.; Guo, L. BnaPPT1 is essential for chloroplast development and seed oil accumulation in Brassica napus. J. Adv. Res. 2022, 42, 29–40. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Ye, S.; Huang, Y.; Ma, T.; Ma, X.; Li, R.; Shen, J.; Wen, J. BnaABF3 and BnaMYB44 regulate the transcription of zeaxanthin epoxidase genes in carotenoid and abscisic acid biosynthesis. Plant Physiol. 2024, 195, 2372–2388. [Google Scholar] [CrossRef] [PubMed]
- Ali, E.; Zhang, K. CRISPR-mediated technology for seed oil improvement in rapeseed: Challenges and future perspectives. Front. Plant Sci. 2023, 14, 1086847. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Raza, A.; Chu, W.; Zou, X.; Cheng, H.; Hu, Q.; Liu, J.; Wei, W. Comprehensive in silico characterization and expression profiling of TCP gene family in rapeseed. Front. Genet. 2021, 12, 794297. [Google Scholar] [CrossRef] [PubMed]
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009, 150, 482–493. [Google Scholar] [CrossRef]
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancón, C.; Immink, R.G.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, E245–E254. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, Y.; Ge, D.; Wang, Z.; Song, W.; Gu, R.; Zhang, X. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efffux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. USA 2019, 116, 17105–17114. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Finlayson, S.A. Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 2015, 169, 611–626. [Google Scholar] [CrossRef]
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Åstot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Korves, T.M.; Bergelson, J. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 2003, 133, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Dong, L.; Wang, Z.; Zhang, H.; Han, C.; Liu, B.; Wang, X.; Chen, Q. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Cardoza, V.; Stewart, C. Increased Agrobacterium-mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyl segment explants. Plant Cell Rep. 2003, 21, 599–604. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Wang, K. HiTOM: A platform for high.throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–DDCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Zhang, J.; Cui, C.; Chai, L.; Zheng, B.; Jiang, L.; Li, H. The WRKY28-BRC1 Transcription Factor Module Controls Shoot Branching in Brassica napus. Plants 2025, 14, 486. https://doi.org/10.3390/plants14030486
Zhang K, Zhang J, Cui C, Chai L, Zheng B, Jiang L, Li H. The WRKY28-BRC1 Transcription Factor Module Controls Shoot Branching in Brassica napus. Plants. 2025; 14(3):486. https://doi.org/10.3390/plants14030486
Chicago/Turabian StyleZhang, Ka, Jinfang Zhang, Cheng Cui, Liang Chai, Benchuan Zheng, Liangcai Jiang, and Haojie Li. 2025. "The WRKY28-BRC1 Transcription Factor Module Controls Shoot Branching in Brassica napus" Plants 14, no. 3: 486. https://doi.org/10.3390/plants14030486
APA StyleZhang, K., Zhang, J., Cui, C., Chai, L., Zheng, B., Jiang, L., & Li, H. (2025). The WRKY28-BRC1 Transcription Factor Module Controls Shoot Branching in Brassica napus. Plants, 14(3), 486. https://doi.org/10.3390/plants14030486





