Trans-Kingdom RNA Dialogues: miRNA and milRNA Networks as Biotechnological Tools for Sustainable Crop Defense and Pathogen Control
Abstract
1. Introduction
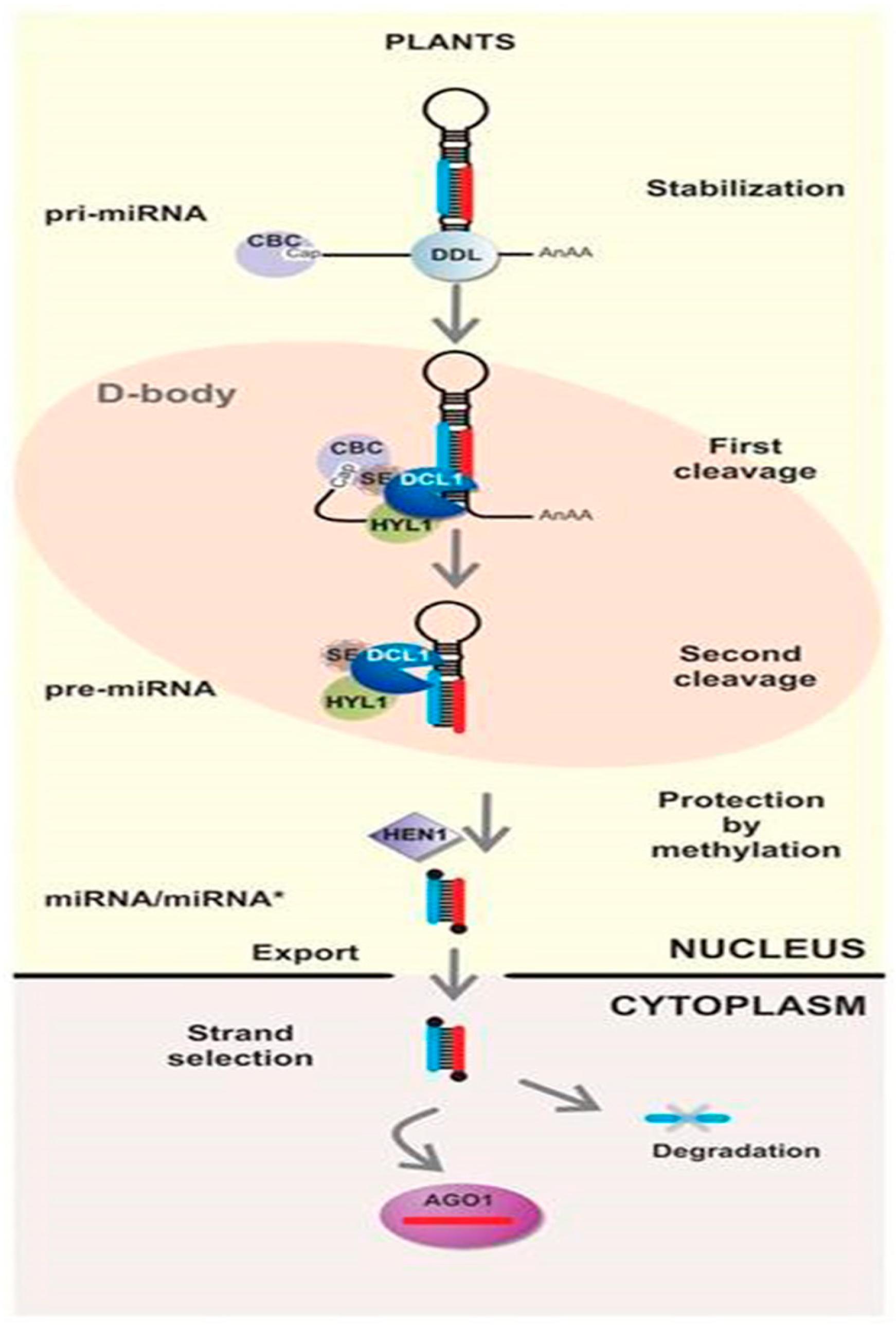
2. miRNA (milRNA) Regulates Endogenous Gene Expression
2.1. miRNAs Regulate Endogenous Gene Expression and Participate in Disease Resistance
2.1.1. miRNAs Regulate PTI
2.1.2. miRNAs Regulate ETI
2.1.3. miRNAs Regulate Hormones Signal Transduction
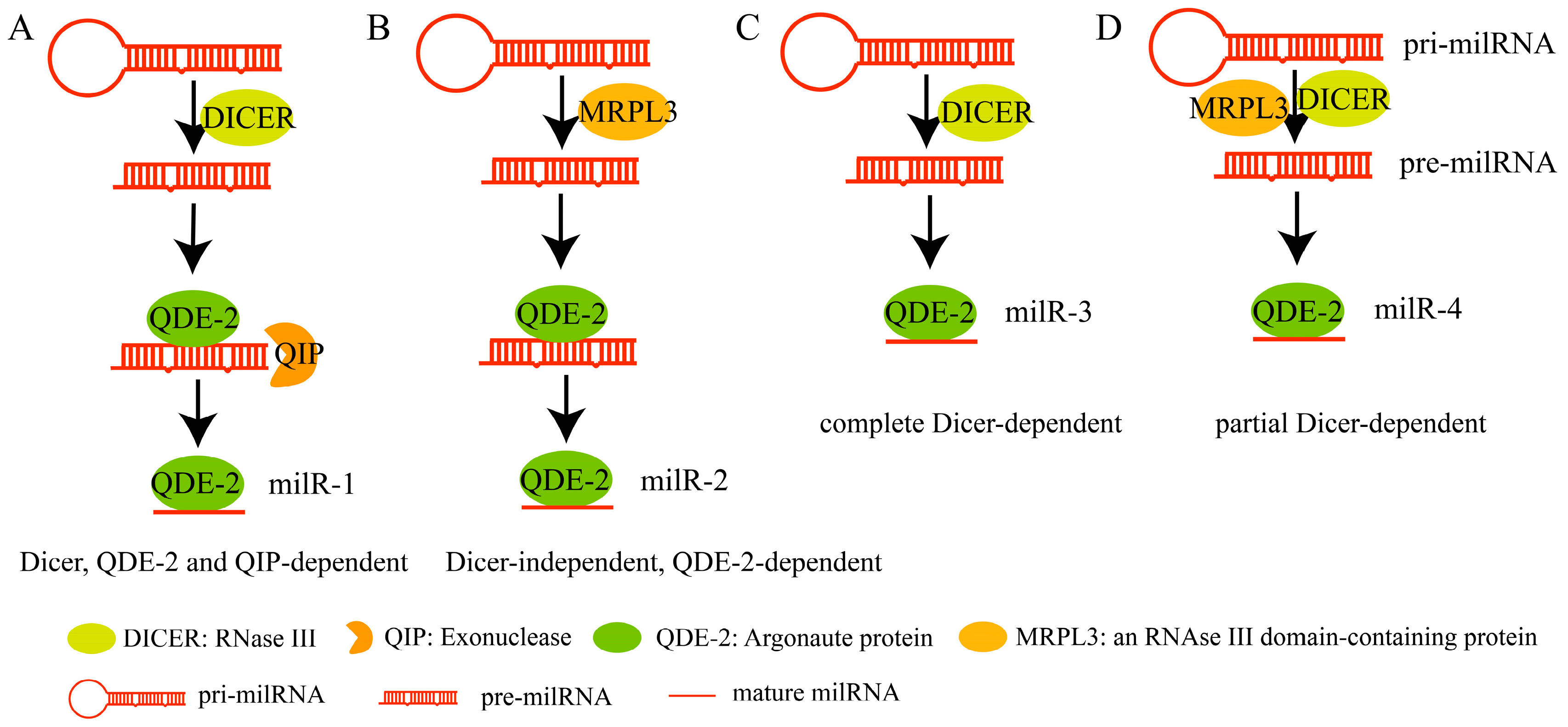
2.2. Fungal milRNAs Regulate Endogenous Gene Expression and Are Involved in Pathogenesis
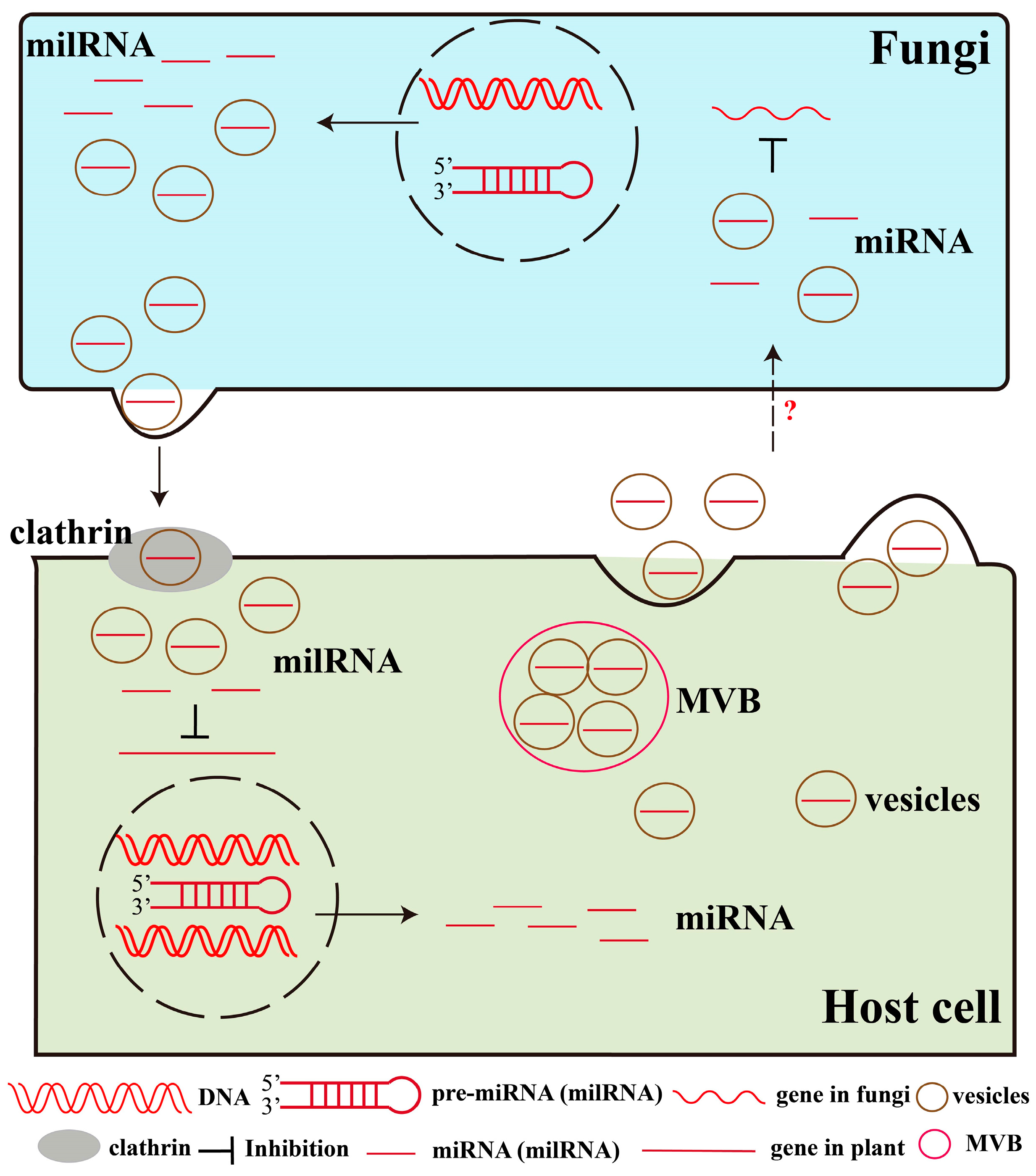
3. miRNAs (milRNAs) Mediate Trans-Kingdom Regulation in Plant–Pathogen Interactions
3.1. Plant miRNAs Modulate Fungal Gene Expression and Suppress Pathogen Invasion
3.2. Fungal milRNAs Reduce Plant Immunity by Regulating Disease-Resistance Gene Expression
3.3. Trans-Kingdom Regulation of miRNAs in the Mycorrhizal Symbiosis
| Host Plant | Pathogen | sRNAs | Target Genes | Directionality of RNA Mobility | Reference |
|---|---|---|---|---|---|
| Zea mays | Fusarium verticillioides | miR528b-5p | FvTTP | Host-pathogen | [35] |
| cotton | Verticillium dahliae | miR166, miR159 | Clp-1, HiC-15 | Host-pathogen | [42] |
| Malus hupehensis | Botryosphaeria dothidea | miR159a | BdSTP | Host-pathogen | [87] |
| Arabidopsis thaliana | Botrytis cinerea | Bc-sRNAs | MPK2, MPK1, PRXIIF, and WAK | Pathogen-host | [91,95] |
| tomato | Fusarium oxysporum | Fol-milR1 | SlyAGO4a | Pathogen-host | [92] |
| banana | Fusarium oxysporum | Foc-milR87 | MaPTI6L | Pathogen-host | [93] |
| banana | Fusarium oxysporum f. sp. cubense | Foc-milR138 | MaLYK3 | Pathogen-host | [94] |
4. Applications of miRNA in Crop Protection and Yield Enhancement
4.1. Biotechnological Applications Based on miRNA Overexpression
4.2. Applications of miRNA Silencing Technology
4.3. CRISPR/Cas9-Mediated Precise miRNA Editing Technology
5. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Presti, L.L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Yu, X.Q.; Niu, H.Q.; Liu, C.; Wang, H.L.; Yin, W.L.; Xia, X.L. PTI-ETI synergistic signal mechanisms in plant immunity. Plant Biotechnol. J. 2024, 22, 2113–2128. [Google Scholar] [CrossRef]
- Yuan, M.H.; Ngou, B.P.M.; Ding, P.T.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X.N. Salicylic acid in Plant immunity and beyond. Plant Cell 2024, 36, 1451–1464. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.T.; Ding, Y.L. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Li, C.; Xu, M.X.; Cai, X.; Han, Z.G.; Si, J.P.; Chen, D.H. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.; Li, Y.J.; Sheng, J.P.; Shen, L. Recent advances in dissecting the function of ethylene in interaction between host and pathogen. J. Agric. Food Chem. 2024, 72, 4552–4563. [Google Scholar] [CrossRef]
- Hou, S.J.; Tsuda, K. Salicylic acid and jasmonic acid crosstalk in plant immunity. Essays Biochem. 2022, 66, 647–656. [Google Scholar] [PubMed]
- Nakano, M.; Omae, N.; Tsuda, K. Inter-organismal phytohormone networks in plant-microbe interactions. Curr. Opin. Plant Biol. 2022, 68, 102258. [Google Scholar] [CrossRef]
- Gim, J.A.; Ha, H.S.; Ahn, K.; Kim, D.S.; Kim, H.S. Genome-wide identification and classification of microRNAs derived from repetitive elements. Genom. Inform. 2014, 12, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.L.; Kuang, Z.; Tao, Y.T.; Wang, H.T.; Wan, M.M.; Hao, C.; Shen, F.; Yang, X.Z.; Li, L. Miniature inverted-repeat transposable elements drive rapid microRNA diversification in angiosperms. Mol. Biol. Evol. 2022, 39, msac224. [Google Scholar] [CrossRef]
- Xu, W.B.; Zhao, L.; Liu, P.; Guo, Q.H.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zhang, S.X.; Guo, X.Q.; Zhang, S.Z.; et al. Intronic microRNA-directed regulation of mitochondrial reactive oxygen species enhances plant stress tolerance in Arabidopsis. New Phytol. 2023, 240, 710–726. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene Lin-4 encodes small RNAs with antisense complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide Let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of Let-7 heterochronic regulatory RNA. Nature. 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.Q.; Chen, M.; Niu, J.R.; Wang, L.; Li, Y.; Fang, X.F.; Li, P.L.; Qi, Y.J. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 2021, 23, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Han, M.H.; Fedoroff, N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 9970–9975. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Tomczyk, K.; Mickiewicz, A.; Sarzynska, J.; Szachniuk, M. Bioinformatics study of structural patterns in plant microRNA precursors. Biomed Res. Int. 2017, 2017, 6783010. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Szachniuk, M. Discovering structural motifs in miRNA precursors from the Viridiplantae kingdom. Molecules 2018, 23, 1367. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yang, Z.Y.; Li, J.J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X.M. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef]
- Bologna, N.G.; Schapire, A.L.; Palatnik, J.F. Processing of plant microRNA precursors. Brief. Funct. Genom. 2013, 12, 37–45. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Lee, H.C.; Li, L.; Gu, W.F.; Xue, Z.H.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.A.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Q.; Huang, M.; Liu, Y.; Liu, Z.; Liu, X.; Ma, Z. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 2015, 5, 12500. [Google Scholar] [CrossRef]
- Hu, W.C.; Luo, H.; Yang, Y.K.; Wang, Q.; Hong, N.; Wang, G.P.; Wang, A.; Wang, L.P. Comprehensive analysis of full genome sequence and Bd-milRNA/target mRNAs to discover the mechanism of hypovirulence in Botryosphaeria dothidea strains on pear infection with BdCV1 and BdPV1. IMA Fungus 2019, 10, 3. [Google Scholar] [CrossRef]
- Kang, K.; Zhong, J.S.; Jiang, L.; Liu, G.; Gou, C.Y.; Wu, Q.; Wang, Y.; Luo, J.; Gou, D. Identification of microRNA-like RNAs in the filamentous fungus Trichoderma Reesei by solexa sequencing. PLoS ONE 2013, 8, e76288. [Google Scholar] [CrossRef] [PubMed]
- Jeseničnik, T.; Štajner, N.; Radišek, S.; Mishra, A.K.; Košmelj, K.; Kunej, U.; Jakše, J. Discovery of microRNA-like small RNAs in pathogenic plant fungus Verticillium nonalfalfae using high-throughput sequencing and qPCR and RLM-RACE validation. Int. J. Mol. Sci. 2022, 23, 900. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, N.; Jiang, Q.Y.; Sun, X.J.; Wang, Y.; Zhang, H.; Hu, Z. Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum. PLoS ONE 2014, 9, e104956. [Google Scholar] [CrossRef]
- Qu, Q.; Liu, N.; Su, Q.F.; Liu, X.F.; Jia, H.; Liu, Y.W.; Sun, M.L.; Cao, Z.Y.; Dong, J.G. MicroRNAs involved in the trans-kingdom gene regulation in the interaction of maize kernels and Fusarium verticillioides. Int. J. Biol. Macromol. 2023, 242, 125046. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.H.; Fu, Y.P.; Xie, J.T.; Li, B.; Jiang, D.H.; Li, G.Q.; Cheng, J.S. Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol. Genet. Genom. 2012, 287, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B.H. MicroRNAs in control of plant development. J. Cell Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Nadarajah, K.; Kumar, I.S. Drought response in rice: The miRNA story. Int. J. Mol. Sci. 2019, 20, 3766. [Google Scholar] [CrossRef]
- Singh, A.; Jain, D.; Pandey, J.; Yadav, M.; Bansal, K.C.; Singh, I.K. Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit. Rev. Biotechnol. 2023, 43, 613–627. [Google Scholar] [CrossRef]
- Zhang, F.Y.; Yang, J.W.; Zhang, N.; Wu, J.H.; Si, H.J. Roles of microRNAs in abiotic stress response and characteristics regulation of plant. Front. Plant Sci. 2022, 13, 919243. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.F.; Song, N.; Zhao, M.X.; Liu, R.; Feng, H.; Wang, X.J.; Kang, Z.S. Puccinia Striiformis f. sp. Tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.L.; Wang, Y.; Li, Y.F.; Sun, P.L.; Jiang, J.Y.; Zhou, H.N.; Liu, J.N.; Wang, S.B. Expression of mosquito miRNAs in entomopathogenic fungus induces pathogen-mediated host RNA interference and increases fungal efficacy. Cell Rep. 2022, 41, 111527. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.Y.; Kogel, K.H.; Jin, H.L. Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Bundó, M.; Val-Torregrosa, B.; Martín-Cardoso, H.; Ribaya, M.; Campos-Soriano, L.; Bach-Pages, M.; Chiou, T.J.; San Segundo, B. Silencing Osa-miR827 via CRISPR/Cas9 protects rice against the blast fungus Magnaporthe oryzae. Plant Mol. Biol. 2024, 114, 105. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.T.; He, X.R.; Zhu, Y.; Feng, Q.; Yang, X.M.; Zhou, X.H.; Li, G.B.; Ji, Y.P.; Zhao, J.H.; et al. Blocking Osa-miR1871 enhances rice resistance against Magnaporthe oryzae and yield. Plant Biotechnol. J. 2022, 20, 646–659. [Google Scholar] [CrossRef]
- Zhang, L.L.; Huang, Y.Y.; Zheng, Y.P.; Liu, X.X.; Zhou, S.X.; Yang, X.M.; Liu, S.L.; Li, Y.; Li, J.L.; Zhao, S.L.; et al. Osa-miR535 targets SQUAMOSA promoter binding protein-like 4 to regulate blast disease resistance in rice. Plant J. 2022, 110, 166–178. [Google Scholar] [CrossRef]
- Jia, Y.F.; Wei, K.; Qin, J.W.; Zhai, W.X.; Li, Q.L.; Li, Y.N. The roles of microRNAs in the regulation of rice-pathogen interactions. Plants 2025, 14, 136. [Google Scholar] [CrossRef]
- de Oliveira Cabral, S.K.; de Freitas, M.B.; Stadnik, M.J.; Kulcheski, F.R. Emerging roles of plant microRNAs during Colletotrichum spp. infection. Planta 2024, 259, 48. [Google Scholar] [CrossRef]
- Wang, S.Q.; Wang, X.H.; Chen, J. Identification of miRNAs involved in maize-induced systemic resistance primed by Trichoderma harzianum T28 against Cochliobolus heterostrophus. J. Fungi 2023, 9, 278. [Google Scholar] [CrossRef]
- Lv, Y.D.; Zhong, Y.; Jiang, B.; Yan, H.X.; Ren, S.; Cheng, C.Z. MicroRNA miR171b positively regulates resistance to Huanglongbing of citrus. Int. J. Mol. Sci. 2023, 24, 5737. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, H.J.; Kwak, K.J.; Lee, K.; Hong, S.W.; Kang, H. MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014, 55, 1660–1668. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, X.L.; Zhu, Y.; Yang, X.M.; Zhang, K.N.; Xiao, Z.Y.; Wang, H.; Zhao, J.H.; Zhang, L.L.; Li, G.B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef]
- Li, J.; Song, Q.; Zuo, Z.F.; Liu, L. MicroRNA398: A master regulator of plant development and stress responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Yu, Y.R.; Fei, S.H.; Chen, Y.X.; Xu, Y.M.; Zhu, Z.J.; He, Y. Overexpression of Sly-miR398b compromises disease resistance against Botrytis cinerea through regulating ROS homeostasis and JA-related defense genes in tomato. Plants 2023, 12, 2572. [Google Scholar] [CrossRef]
- Xu, W.; Meng, Y.; Wise, R.P. Mla- and Rom1-mediated control of microRNA398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytol. 2014, 201, 1396–1412. [Google Scholar] [CrossRef]
- Salvador-Guirao, R.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol. Plant Microbe Interact. 2018, 31, 249–259. [Google Scholar] [CrossRef]
- Wang, J.L.; Hu, T.H.; Wang, W.H.; Hu, H.J.; Wei, Q.Z.; Bao, C.L. Investigation of evolutionary and expressional relationships in the function of the leucine-rich repeat receptor-like protein kinase gene family (LRR-RLK) in the radish (Raphanus sativus L.). Sci. Rep. 2019, 9, 6937. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. The role of plant small RNAs in NB-LRR regulation. Brief. Funct. Genom. 2015, 14, 268–274. [Google Scholar] [CrossRef]
- Yang, L.; Mu, X.Y.; Liu, C.; Cai, J.H.; Shi, K.; Zhu, W.J.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef]
- Song, X.W.; Li, P.C.; Zhai, J.X.; Zhou, M.; Ma, L.J.; Liu, B.; Jeong, D.H.; Nakano, M.; Cao, S.Y.; Liu, C.Y.; et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. Cell Mol. Biol. 2012, 69, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Y.; Li, H.G.; Wang, Y.L.; Li, S.; Wang, H.L.; Yu, L.; He, F.; Yang, Y.; Feng, C.H.; Shuai, P.; et al. Poplar miR472a targeting NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma. J. Exp. Bot. 2018, 69, 5519–5530. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Fan, L.J.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef] [PubMed]
- Różańska, E.; Krępski, T.; Wiśniewska, A. Mutations in selected ABA-related genes reduce level of Arabidopsis thaliana susceptibility to the beet cyst nematode Heterodera schachtii. Plants 2023, 12, 2299. [Google Scholar] [CrossRef]
- Zhu, T.; Zhou, X.; Zhang, J.L.; Zhang, W.H.; Zhang, L.P.; You, C.X.; Jameson, P.E.; Ma, P.T.; Guo, S.L. Ethylene-induced NbMYB4L is involved in resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Pathol. 2022, 23, 16–31. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef]
- Palmer, I.A.; Chen, H.; Chen, J.; Chang, M.; Li, M.; Liu, F.Q.; Fu, Z.Q. Novel salicylic acid analogs induce a potent defense response in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 3356. [Google Scholar] [CrossRef]
- Truman, W.; Sreekanta, S.; Lu, Y.; Bethke, G.; Tsuda, K.; Katagiri, F.; Glazebrook, J. The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 2013, 163, 1741–1751. [Google Scholar] [CrossRef]
- Hu, G.; Hao, M.Y.; Wang, L.; Liu, J.F.; Zhang, Z.N.; Tang, Y.; Peng, Q.Z.; Yang, Z.R.; Wu, J.H. The cotton miR477-CBP60A module participates in plant defense against Verticillium dahlia. Mol. Plant Microbe Interact. 2020, 33, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Wang, R.J.; Ma, P.P.; Cao, J.S.; Cao, Y.; Zhou, Z.J.; Li, T.; Wu, J.Y.; Zhang, H.Y. A novel maize microRNA negatively regulates resistance to Fusarium verticillioides. Mol. Plant Pathol. 2022, 23, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Xu, M.; Gao, Y.Q.; Liang, J.H.; Guo, F.R.; Guo, Y.; Huang, L.L. Vm-milR37 contributes to pathogenicity by regulating glutathione peroxidase gene VmGP in Valsa mali. Mol. Plant Pathol. 2021, 22, 243–254. [Google Scholar] [CrossRef]
- Xu, M.; Gao, C.; Ji, L.; Zhu, L.H.; Gao, Y.Q.; Feng, H.; Huang, L.L. A Fungal microRNA-like RNA regulated effector promotes pathogen infection by targeting a host defense-related transcription factor. Plant J. 2023, 115, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Guo, Y.; Tian, R.Z.; Gao, C.; Guo, F.R.; Voegele, R.T.; Bao, J.Y.; Li, C.J.; Jia, C.H.; Feng, H.; et al. Adaptive regulation of virulence genes by microRNA-like RNAs in Valsa mali. New Phytol. 2020, 227, 899–913. [Google Scholar] [CrossRef]
- Li, M.H.; Xie, L.F.; Wang, M.; Lin, Y.L.; Zhong, J.Q.; Zhang, Y.; Zeng, J.; Kong, G.H.; Xi, P.P.; Li, H.P.; et al. FoQDE2-dependent milRNA promotes Fusarium oxysporum f. sp. cubense virulence by silencing a glycosyl hydrolase coding gene expression. PLoS Pathog. 2022, 18, e1010157. [Google Scholar] [CrossRef]
- Mathur, M.; Nair, A.; Kadoo, N. Plant-pathogen interactions: microRNA-mediated trans-kingdom gene regulation in fungi and their host plants. Genomics 2020, 112, 3021–3035. [Google Scholar] [CrossRef]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-kingdom RNA silencing in plant-fungal pathogen interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef]
- Zhao, J.H.; Guo, H.S. Trans-kingdom RNA interactions drive the evolutionary arms race between hosts and pathogens. Curr. Opin. Genet. Dev. 2019, 58–59, 62–69. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.J.; Zhao, J.H.; Fang, Y.Y.; He, X.F.; Guo, H.S.; Duan, C.G. A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef]
- Raruang, Y.; Omolehin, O.; Hu, D.; Wei, Q.J.; Promyou, S.; Parekattil, L.J.; Rajasekaran, K.; Cary, J.W.; Wang, K.; Chen, Z.Y. Targeting the Aspergillus flavus P2c gene through host-Induced gene silencing reduces A. flavus infection and aflatoxin contamination in transgenic maize. Front. Plant Sci. 2023, 14, 1150086. [Google Scholar] [CrossRef]
- Guo, J.; Mou, Y.; Li, Y.X.; Yang, Q.; Wang, X.; Lin, H.C.; Kang, Z.S.; Guo, J. Silencing a chitinase gene, PstChia1, reduces virulence of Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2023, 24, 8215. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Yogendra, K.; Sanivarapu, H.; Rajasekaran, K.; Cary, J.W.; Sharma, K.K.; Bhatnagar-Mathur, P. Multiplexed host-induced gene silencing of Aspergillus flavus genes confers aflatoxin resistance in groundnut. Toxins 2023, 15, 319. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.L.; Wang, M.; He, B.Y.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H.L. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence gGenes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Zhou, Q.F.; Ma, K.; Hu, H.H.; Xing, X.L.; Huang, X.; Gao, H. Extracellular vesicles: Their functions in plant-pathogen interactions. Mol. Plant Pathol. 2022, 23, 760–771. [Google Scholar] [CrossRef]
- Krylova, S.V.; Feng, D.R. The machinery of exosomes: Biogenesis, release, and uptake. Int. J. Mol. Sci. 2023, 24, 1337. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Y.; Lin, X.X.; Zhou, T.T.; Cao, L.F.; Hu, K.X.; Li, F.Z.; Qu, S.C. Host-induced gene silencing in wild apple germplasm malus hupehensis confers resistance to the fungal pathogen Botryosphaeria dothidea. Plant J. 2024, 118, 1174–1193. [Google Scholar] [CrossRef]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.-Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H.L. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef]
- Wu, Q.L.; Liu, P.; Liu, X.L.; Li, G.Q.; Huang, L.; Ying, F.Q.; Gong, L.Q.; Li, W.H.; Zhang, J.N.; Gao, R.; et al. hnRNPA2B1 facilitates ovarian carcinoma metastasis by sorting cargoes into small extracellular vesicles driving myofibroblasts activation. J. Nanobiotechnol. 2025, 23, 273. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.M.; Mao, H.Y.; Li, S.J.; Feng, T.; Zhang, Z.Y.; Cheng, L.; Luo, S.J.; Borkovich, K.A.; Ouyang, S.Q. Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol. 2021, 232, 705–718. [Google Scholar] [CrossRef]
- Zhong, J.Q.; Situ, J.J.; He, C.C.; He, J.H.; Kong, G.H.; Li, H.P.; Jiang, Z.D.; Li, M.H. A virulent milRNA of Fusarium oxysporum f. sp. cubense impairs plant resistance by targeting banana AP2 transcription factor coding gene MaPTI6L. Hortic. Res. 2024, 12, uhae361. [Google Scholar] [CrossRef] [PubMed]
- He, J.H.; Zhong, J.Q.; Jin, L.Q.; Long, Y.K.; Situ, J.J.; He, C.C.; Kong, G.H.; Jiang, Z.D.; Li, M.H. A virulent milRNA inhibits host immunity by silencing a host receptor-like kinase MaLYK3 and facilitates infection by Fusarium oxysporum f. sp. cubense. Mol. Plant Pathol. 2024, 25, e70016. [Google Scholar] [CrossRef] [PubMed]
- He, B.H.; Wang, H.; Liu, G.S.; Chen, A.; Calvo, A.; Cai, Q.; Jin, H.L. Fungal small RNAs ride in extracellular vesicles to enter plant cells through clathrin-mediated endocytosis. Nat. Commun. 2023, 14, 4383. [Google Scholar] [CrossRef]
- Ren, B.; Wang, X.T.; Duan, J.B.; Ma, J.X. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef]
- Silvestri, A.; Turina, M.; Fiorilli, V.; Miozzi, L.; Venice, F.; Bonfante, P.; Lanfranco, L. Different genetic sources contribute to the small RNA population in the arbuscular mycorrhizal fungus Gigaspora margarita. Front. Microbiol. 2020, 11, 395. [Google Scholar] [CrossRef]
- Wong-Bajracharya, J.; Singan, V.R.; Monti, R.; Plett, K.L.; Ng, V.; Grigoriev, I.V.; Martin, F.M.; Anderson, I.C.; Plett, J.M. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2103527119. [Google Scholar] [CrossRef]
- Lee, J.H.; Won, H.J.; Oh, E.S.; Oh, M.H.; Jung, J.H. Golden gate cloning-compatible DNA replicon/2A-mediated polycistronic vectors for plants. Front. Plant Sci. 2020, 11, 559365. [Google Scholar] [CrossRef]
- Tang, G.L.; Yan, J.; Gu, Y.Y.; Qiao, M.M.; Fan, R.W.; Mao, Y.P.; Tang, X.Q. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.S.; Yan, J.; Gou, F.; Mao, Y.F.; Tang, G.L.; Botella, J.R.; Zhu, J.K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed]
- Lukan, T.; Veillet, F.; Križnik, M.; Coll, A.; Mahkovec Povalej, T.; Pogačar, K.; Stare, K.; Chauvin, L.; Chauvin, J.E.; Gruden, K. CRISPR/Cas9-mediated fine-tuning of miRNA expression in tetraploid potato. Hortic. Res. 2022, 9, uhac147. [Google Scholar] [CrossRef]
- Charpentier, E.; Richter, H.; van der Oost, J.; White, M.F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 2015, 39, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Wang, C.; Sheng, W.J.; Zhou, Y.; Hang, X.D.; Zhao, J.Y.; Gu, Y.Y.; Meng, X.F.; Bai, Y.F.; Li, W.L.; Zhang, Y.J.; et al. siRNA-AGO2 complex inhibits bacterial gene translation: A promising therapeutic strategy for superbug infection. Cell Rep. Med. 2025, 6, 101997. [Google Scholar] [CrossRef]
- Cao, W.D.; Huang, Y.C.; Xu, T.L. RNAi-A new approach to combat bacterial drug resistance. J. Agric. Sci. 2024, 45, 79–83. [Google Scholar]
- Tian, W.; Zhang, T.; Zhao, J.H.; Dong, Y.M.; Li, Y.Z.; Zhao, Z.Q.; Gao, F.; Wu, X.M.; Zhang, B.S.; Fang, Y.Y.; et al. HIGS-mediated crop protection against cotton aphids. Plant Biotechnol. J. 2025, 23, 692–694. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria Graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef]
- Pliego, C.; Nowara, D.; Bonciani, G.; Gheorghe, D.M.; Xu, R.; Surana, P.; Whigham, E.; Nettleton, D.; Bogdanove, A.J.; Wise, R.P.; et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 2013, 26, 633–642. [Google Scholar] [CrossRef]
- Wang, M.; Dean, R.A. Host induced gene silencing of Magnaporthe Oryzae by targeting pathogenicity and development genes to control rice blast disease. Front. Plant Sci. 2022, 13, 959641. [Google Scholar] [CrossRef]
- Wu, H.M.; Qiu, J.F.; Zhang, P.R.; Lu, S.; Meng, J.R.; Huang, X.C.; Li, R.; Chen, B.S. Host-induced gene silencing of Sporisorium scitamineum enhances resistance to smut in sugarcane. Plant Biotechnol. J. 2025, 23, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.P.; Wang, J.; Wang, Y.; Lin, J.; Fu, Y.P.; Xie, J.T.; Jiang, D.H.; Chen, T.; Liu, H.Q.; Cheng, J.S. Dicer-like proteins regulate sexual development via the biogenesis of perithecium-specific microRNAs in a plant pathogenic fungus Fusarium graminearum. Front. Microbiol. 2018, 9, 818. [Google Scholar] [CrossRef]
- Mosquera, S.; Ginésy, M.; Bocos-Asenjo, I.T.; Amin, H.; Diez-Hermano, S.; Diez, J.J.; Niño-Sánchez, J. Spray-induced gene silencing to control plant pathogenic fungi: A step-by-step guide. J. Integr. Plant Biol. 2025, 67, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.N.; Hamby, R.; Heller, J.; Zhao, H.W.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- McRae, A.G.; Taneja, J.; Yee, K.; Shi, X.; Haridas, S.; LaButti, K.; Singan, V.; Grigoriev, I.V.; Wildermuth, M.C. Spray-induced gene silencing to identify powdery mildew gene targets and processes for powdery mildew control. Mol. Plant Pathol. 2023, 24, 1168–1183. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H.L. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef]
- Bocos-Asenjo, I.T.; Amin, H.; Mosquera, S.; Díez-Hermano, S.; Ginésy, M.; Diez, J.J.; Niño-Sánchez, J. Spray-induced gene silencing (SIGS) as a tool for the management of pine pitch canker forest disease. Plant Dis. 2025, 109, 49–62. [Google Scholar] [CrossRef]
- Ma, Z.F.; Wang, J.Y.; Li, C.Y. Research progress on miRNAs and artificial miRNAs in insect and disease resistance and breeding in plants. Genes 2024, 15, 1200. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.L.; Zhu, C.H.; Yi, S.J.; Zhang, Y.; Hong, Z. Plant-derived artificial miRNA effectively Reduced the proliferation of aphid (Aphidoidea) through spray-induced gene silencing. Pest Manag. Sci. 2024, 80, 4322–4332. [Google Scholar] [CrossRef]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef]
- Mickiewicz, A.; Rybarczyk, A.; Sarzynska, J.; Figlerowicz, M.; Blazewicz, J. AmiRNA Designer -New method of artificial miRNA design. Acta Biochim. Pol. 2016, 63, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Dumontel, B.; Jiménez-Jiménez, C.; Vallet-Regí, M.; Manzano, M. Bioinspired extracellular vesicle-coated silica nanoparticles as selective delivery systems. Mater. Today Bio 2023, 23, 100850. [Google Scholar] [CrossRef] [PubMed]
- Matta, J.; Maalouf, R. Delivery of siRNA therapeutics: PLGA nanoparticles approach. Front Biosci. Sch. Ed. 2019, 11, 56–74. [Google Scholar]
- Li, Y.; Lu, Y.G.; Shi, Y.; Wu, L.; Xu, Y.J.; Huang, F.; Guo, X.Y.; Zhang, Y.; Fan, J.; Zhao, J.Q.; et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef]
- Yue, E.; Liu, Z.; Li, C.; Li, Y.; Liu, Q.X.; Xu, J.H. Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.). Plant Cell Rep. 2017, 36, 1171–1182. [Google Scholar] [CrossRef]
- Luo, Y.Y.; Wang, L.X.; Zhu, J.; Tian, J.W.; You, L.; Luo, Q.; Li, J.; Yao, Q.; Duan, D. The grapevine miR827a regulates the synthesis of stilbenes by targeting VqMYB14 and gives rise to susceptibility in plant immunity. Theor. Appl. Genet. 2024, 137, 95. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Li, P.; Li, M.; Liu, N.; Dong, J.; Qu, Q.; Cao, Z. Trans-Kingdom RNA Dialogues: miRNA and milRNA Networks as Biotechnological Tools for Sustainable Crop Defense and Pathogen Control. Plants 2025, 14, 1250. https://doi.org/10.3390/plants14081250
Jia H, Li P, Li M, Liu N, Dong J, Qu Q, Cao Z. Trans-Kingdom RNA Dialogues: miRNA and milRNA Networks as Biotechnological Tools for Sustainable Crop Defense and Pathogen Control. Plants. 2025; 14(8):1250. https://doi.org/10.3390/plants14081250
Chicago/Turabian StyleJia, Hui, Pan Li, Minye Li, Ning Liu, Jingao Dong, Qing Qu, and Zhiyan Cao. 2025. "Trans-Kingdom RNA Dialogues: miRNA and milRNA Networks as Biotechnological Tools for Sustainable Crop Defense and Pathogen Control" Plants 14, no. 8: 1250. https://doi.org/10.3390/plants14081250
APA StyleJia, H., Li, P., Li, M., Liu, N., Dong, J., Qu, Q., & Cao, Z. (2025). Trans-Kingdom RNA Dialogues: miRNA and milRNA Networks as Biotechnological Tools for Sustainable Crop Defense and Pathogen Control. Plants, 14(8), 1250. https://doi.org/10.3390/plants14081250





