First Report of a Psyllid Vector of ‘Candidatus Phytoplasma pruni’ (Strain 16SrIII-J)
Abstract
:1. Introduction
2. Results
2.1. Psyllid Collection and Morphological Identification
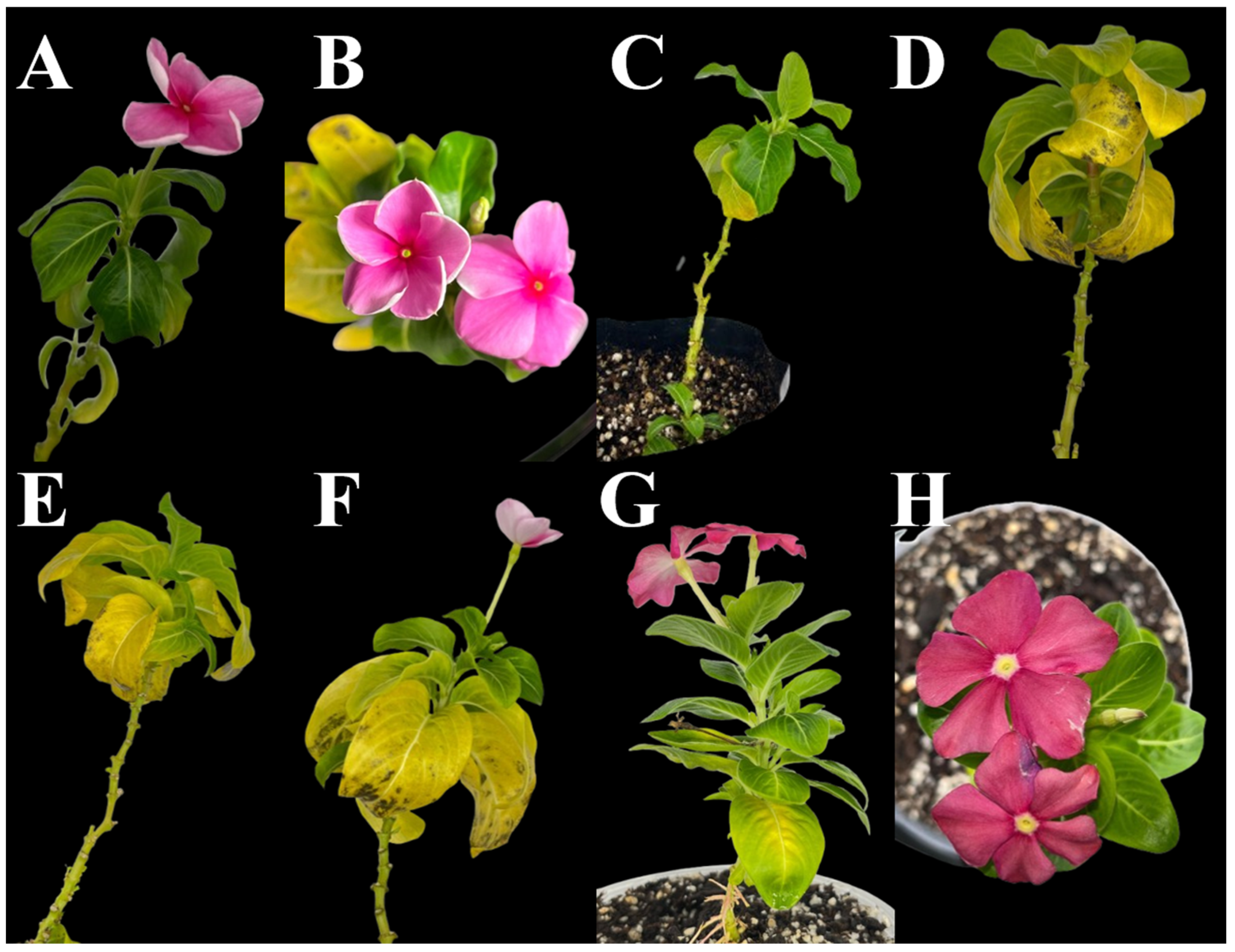
2.2. Transmission Trials
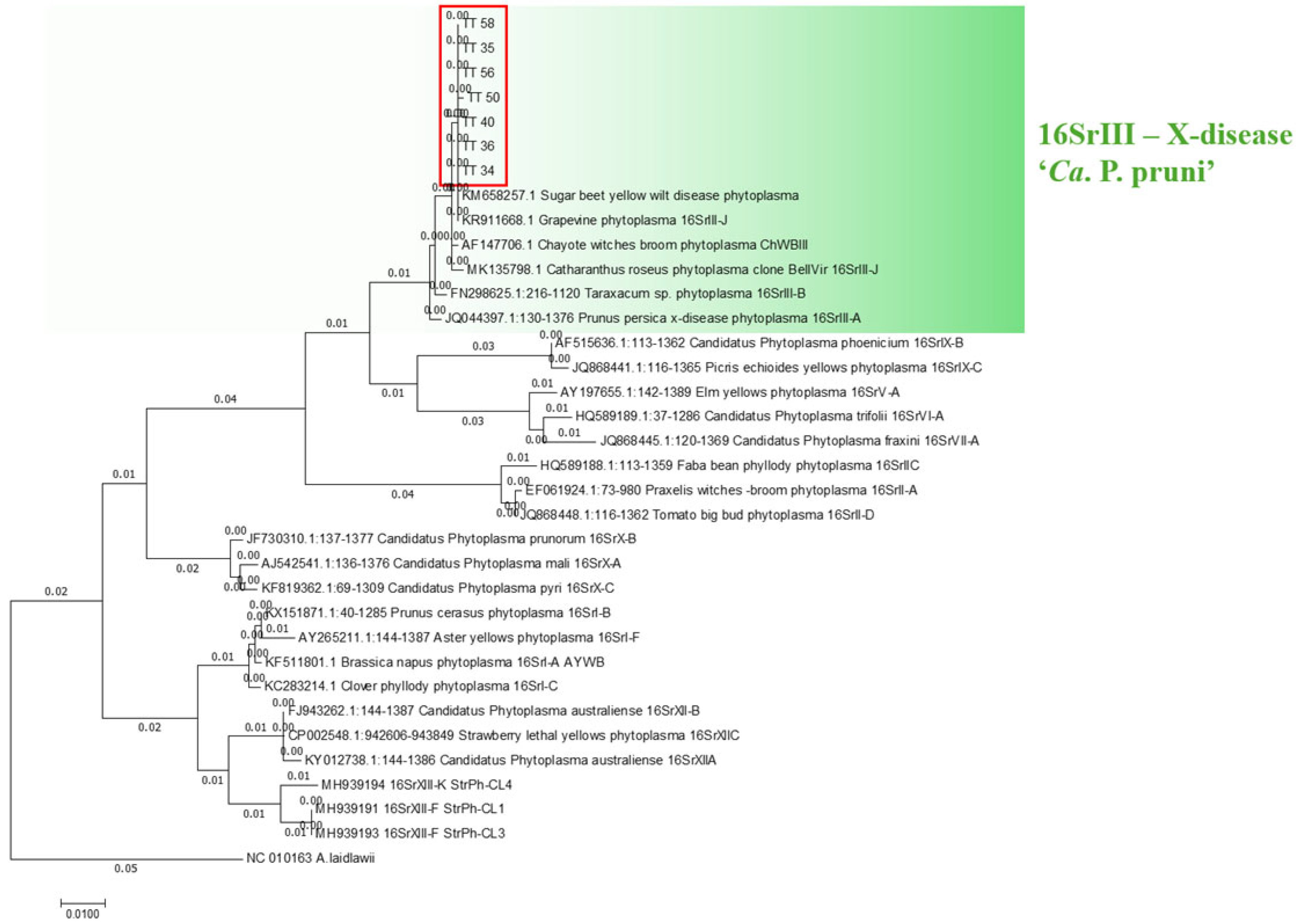
2.3. Phytoplasma Detection
2.4. Detection of the SAP54 Effector
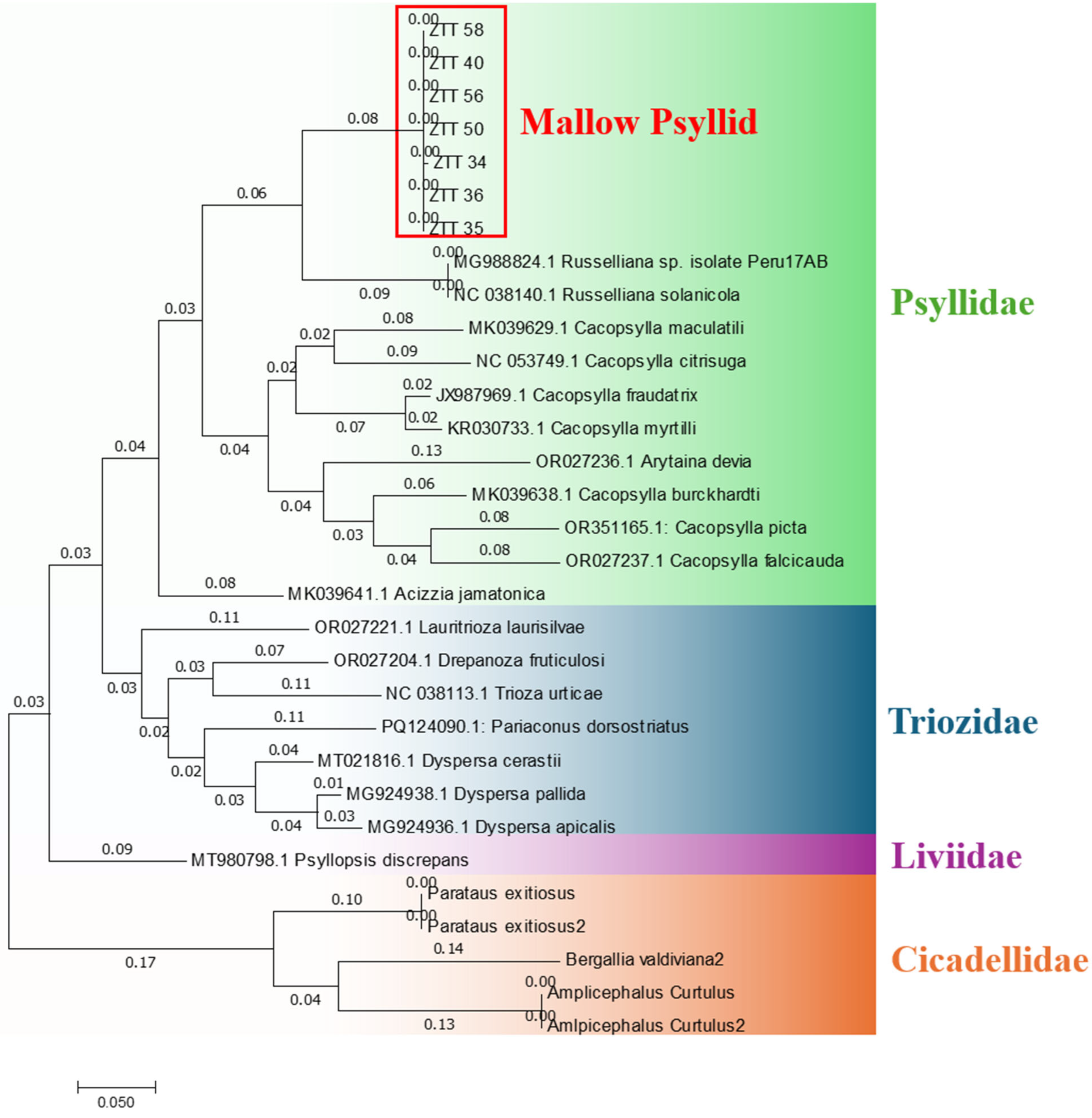
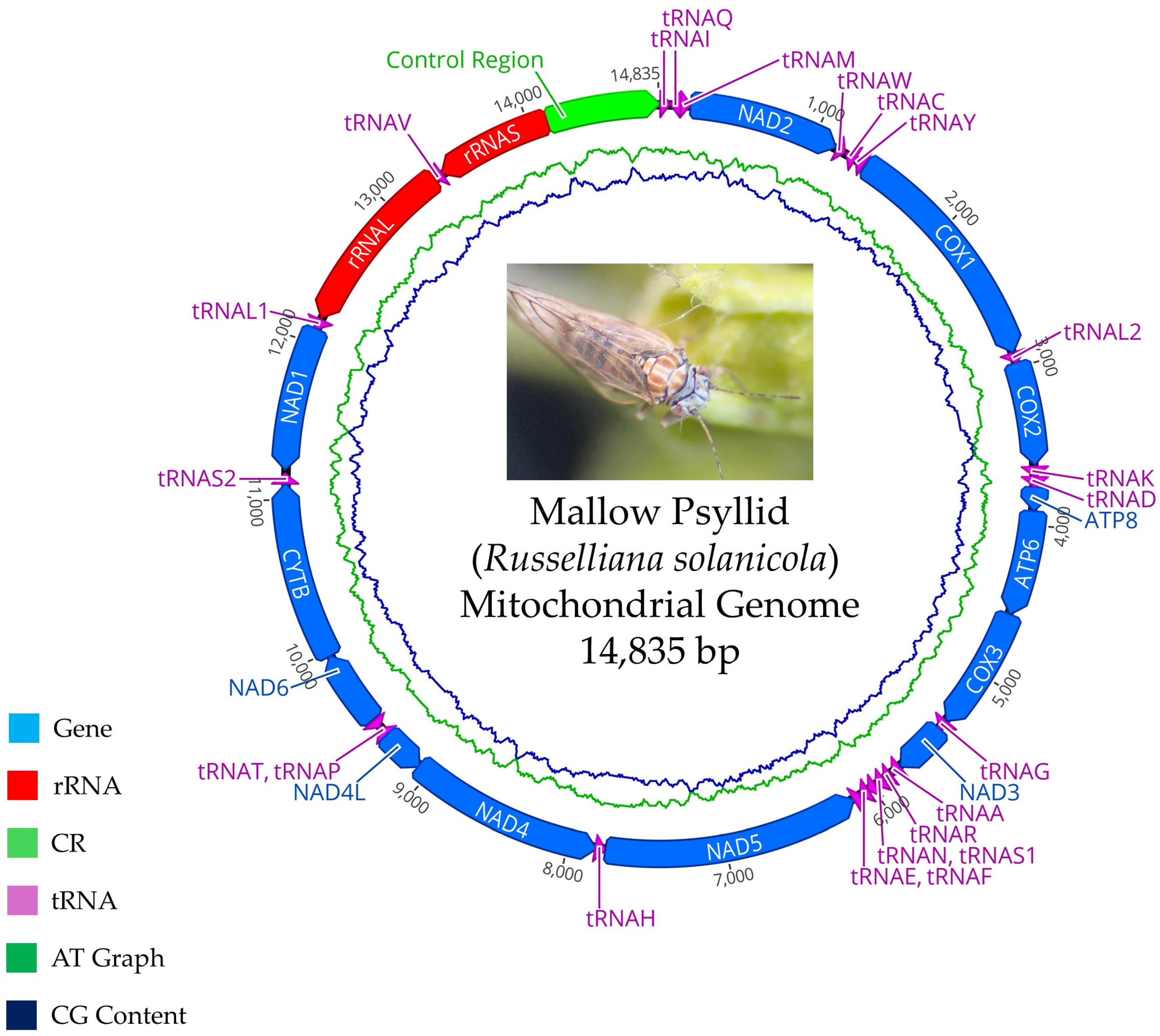
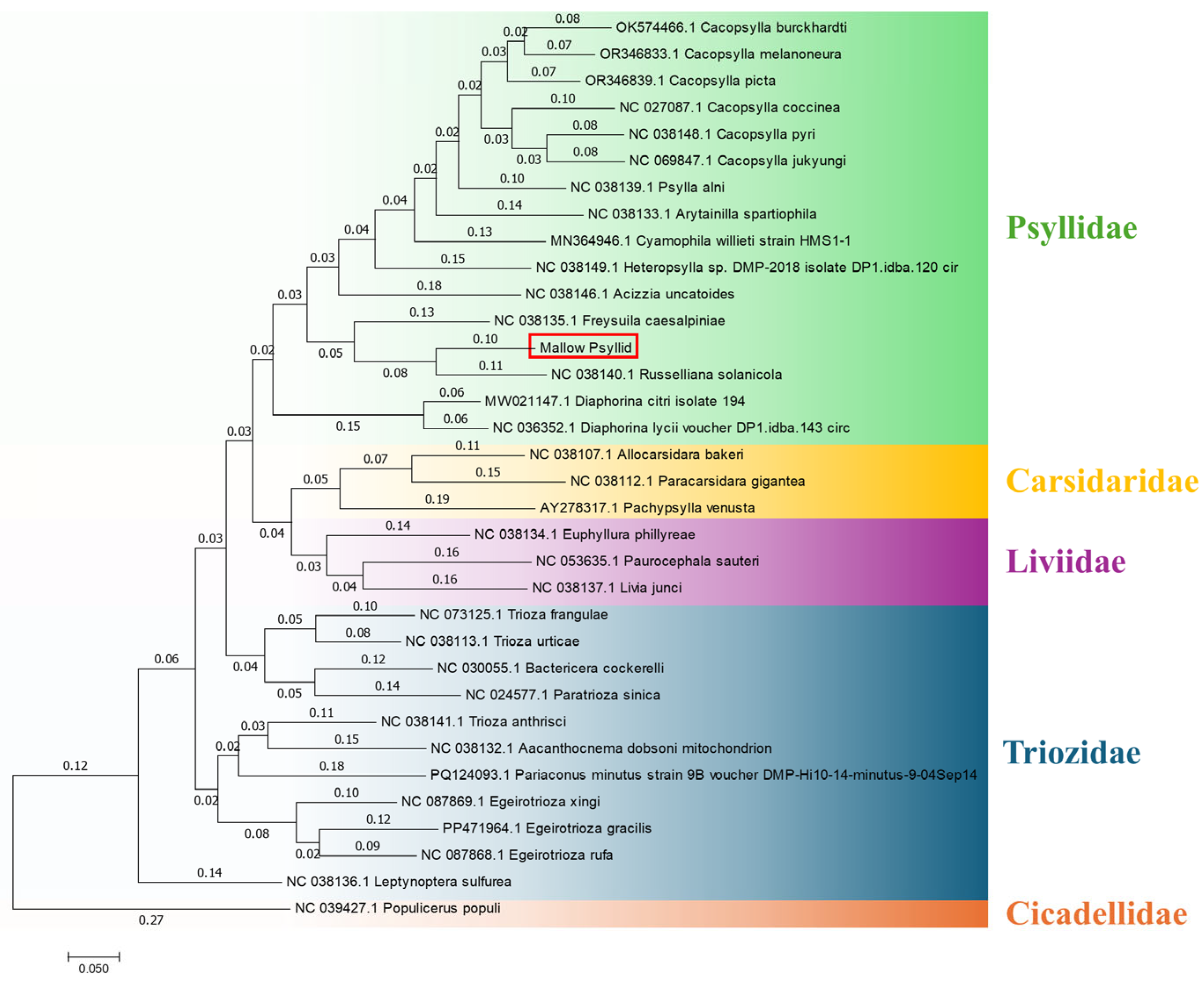
2.5. Molecular Identification of Psyllids
3. Discussion
4. Materials and Methods
4.1. Psyllid Collection
4.2. Psyllid Morphological Identification
4.3. Transmission Trials
4.4. DNA Extraction from Plants Used in Transmission Trials
4.5. DNA Extraction from Psyllids
4.6. Molecular Detection of Phytoplasmas in Psyllids and Plants Used in Transmission Trials
4.7. Detection of the SAP54 Effector Associated with Virescence and Phyllody
4.8. COI Gene Amplification
4.9. Mitochondrial Genome Characterization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Firrao, G.; Gibb, K.; Streten, C. Short Taxonomic Guide to the Genus ‘Candidatus Phytoplasma’. J. Plant Pathol. 2005, 87, 249–263. [Google Scholar]
- Pace, N.; Stahl, D.; Lane, D.; Olsen, G. The Analysis of Natural Microbial Populations by Ribosomal RNA Sequences. In Advances in Microbial Ecology; Springer: Berlin/Heidelberg, Germany, 1986; 55p. [Google Scholar]
- Valenzuela-González, F.; Casillas-Hernández, R.; Villalpando, E.; Vargas-Albores, F. The 16S rRNA Gene in the Study of Marine Microbial Communities. Cienc. Mar. 2015, 14, 297–313. [Google Scholar] [CrossRef]
- Marcone, C. Molecular Biology and Pathogenicity of Phytoplasmas. Ann. Appl. Biol. 2014, 165, 199–221. [Google Scholar] [CrossRef]
- Uribe-Álvarez, C.; Chiquete, N. Las Enfermedades Transmitidas por Vectores y el Potencial Uso de Wolbachia, una Bacteria Endocelular Obligada, para Erradicarlas. Rev. Fac. Med. 2017, 60, 51–55. [Google Scholar]
- Burckhardt, D.; Ouvrard, D.; Percy, D.M. An updated classification of the jumping plant-lice (Hemiptera: Psylloidea) integrating molecular and morphological evidence. Eur. J. Taxon. 2021, 736, 137–182. [Google Scholar] [CrossRef]
- Burckhardt, D.; Ouvrard, D.; Queiroz, D.L.; Percy, D.M. Psyllid Host-Plants (Hemiptera: Psylloidea): Resolving a Semantic Problem. Fla. Entomol. 2014, 97, 242–246. [Google Scholar] [CrossRef]
- Ouvrard, D.; Chalise, P.; Percy, D.M. Host-Plant Leaps Versus Host-Plant Shuffle: A Global Survey Reveals Contrasting Patterns in an Oligophagous Insect Group (Hemiptera, Psylloidea). Syst. Biodivers. 2015, 13, 434–454. [Google Scholar] [CrossRef]
- Serbina, L.; Burckhardt, D. Systematics, biogeography and host-plant relationships of the Neotropical jumping plant-louse genus Russelliana (Hemiptera: Psylloidea). Zootaxa 2017, 4266, 1–114. [Google Scholar] [CrossRef]
- Burckhardt, D.; Queiroz, D.L. Thought to Be Extinct, but Still Alive Today: The Miocene Genus Primascena Klimaszewski, 1997 (Hemiptera: Psyllidae) in the Light of Two Extant Species from Brazil. Insects 2024, 15, 382. [Google Scholar] [CrossRef]
- Serbina, L.; Burckhardt, D.; Birkhofer, K.; Syfert, M.M.; Halbert, S.E. The potato pest Russelliana solanicola Tuthill (Hemiptera: Psylloidea): Taxonomy and host-plant patterns. Zootaxa 2015, 4021, 33–62. [Google Scholar] [CrossRef]
- Bertaccini, A.; Duduk, B. Phytoplasma and Phytoplasma Diseases: A Review of Recent Research. Phytopathol. Mediterr. 2009, 48, 355–378. [Google Scholar]
- Alma, A.; Lessio, F.; Nickel, H. Insects as Phytoplasma Vectors: Ecological and Epidemiological Aspects. In Phytoplasmas: Plant Pathogenic Bacteria—II: Transmission and Management of Phytoplasma-Associated Diseases; Bertaccini, A., Weintraub, P.G., Rao, G.P., Mori, N., Eds.; Gateway East: Singapore, 2019; pp. 1–25. [Google Scholar]
- Navrátil, M.; Válová, P.; Fialová, R.; Lauterer, P.; Šafářová, D.; Starý, M. The Incidence of Stolbur Disease and Associated Yield Losses in Vegetable Crops in South Moravia. Crop Prot. 2009, 28, 898–904. [Google Scholar] [CrossRef]
- Longone, V.; González, F.; Zamorano, A.; Pino, A.M.; Araya, J.; Díaz, V.; Fiore, N. Epidemiological Aspects of Phytoplasmas in Chilean Grapevines. Bull. Insectol. 2011, 64, 85–94. [Google Scholar]
- Quiroga, N.; Gamboa, C.; Soto, D.; Pino, A.M.; Zamorano, A.; Campodonico, J.; Fiore, N. Update and New Epidemiological Aspects about Grapevine Yellows in Chile. Pathogens 2020, 9, 933. [Google Scholar] [CrossRef] [PubMed]
- Camarena Gutiérrez, G.; Torre Almaraz, R. Fitoplasmas: Síntomas y Características Moleculares. Rev. Chapingo Ser. Cienc. For. Ambient. 2008, 14, 81–87. [Google Scholar]
- Kumari, S.; Nagendran, K.; Rai, A.B.; Singh, B.; Rao, G.P.; Bertaccini, A. Global Status of Phytoplasma Diseases in Vegetable Crops. Front. Microbiol. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Facundo, R.; Quiroga, N.; Méndez, P.; Zamorano, A.; Fiore, N. First Report of ‘Candidatus Phytoplasma pyri’ on Pear in Chile. Plant Dis. 2017, 101, 830. [Google Scholar] [CrossRef]
- Gajardo, A.; Fiore, N.; Prodan, S.; Paltrinieri, S.; Botti, S.; Pino, A.M.; Zamorano, A.; Montealegre, J.; Bertaccini, A. Phytoplasmas Associated with Grapevine Yellows Disease in Chile. Plant Dis. 2009, 93, 789–796. [Google Scholar] [CrossRef]
- Fiore, N.; Zamorano, A.; Pino, A. Identification of Phytoplasmas Belonging to the Ribosomal Groups 16SrIII and 16SrV in Chilean Grapevines. Phytopathog. Mollicutes 2015, 5, 32–36. [Google Scholar] [CrossRef]
- Quiroga, N.; Bustamante, M.; Gamboa, C.; Molina, J.; Zamorano, A.; Fiore, N. 16SrIII-J Phytoplasmas Infecting Lettuce and Swiss Chard Crops in Chile. Phytopathog. Mollicutes 2017, 7, 91–94. [Google Scholar] [CrossRef]
- Quiroga, N.; Gamboa, C.; Medina, G.; Contaldo, N.; Torres, F.; Bertaccini, A.; Fiore, N. Survey for ‘Candidatus Liberibacter’ and ‘Candidatus Phytoplasma’ in Citrus in Chile. Pathogens 2022, 11, 1187. [Google Scholar] [CrossRef]
- Cui, W.; Quiroga, N.; Curkovic, T.; Zamorano, A.; Fiore, N. Detection and Identification of 16SrXIII-F and a Novel 16SrXIII Phytoplasma Subgroups Associated with Strawberry Phyllody in Chile. Eur. J. Plant Pathol. 2019, 155, 1039–1046. [Google Scholar] [CrossRef]
- Fuentes, J. Aspectos Epidemiológicos del Decaimiento del Peral en Chile. In Proceedings of the 30th Congress of the Chilean Society of Phytopathology (30° Congreso SOCHIFIT), Santa Cruz, Chile, 13–15 December 2023. [Google Scholar]
- Quiroga, N.; Longone, V.; González, X.; Zamorano, A.; Pino, A.M.; Picciau, L.; Alma, A.; Paltrinieri, S.; Contaldo, N.; Bertaccini, A.; et al. Transmission of 16SrIII-J Phytoplasmas by the Leafhoppers Paratanus exitiosus and Bergallia valdiviana. Phytopathol. Mediterr. 2019, 58, 231–237. [Google Scholar]
- Quiroga, N.; Medina, G.; Zamorano, A.; Acuña, I.; Piña, R.; Fiore, N. New Diseases Associated with 16SrIII-J Phytoplasmas in Chile. Phytopathog. Mollicutes 2019, 9, 15–16. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef]
- Fernández, F.; Debat, H.; Conci, L. Molecular Characterization of Effector Protein SAP54 in Bellis Virescence Phytoplasma (16SrIII-J). Trop. Plant Pathol. 2018, 44, 392–397. [Google Scholar] [CrossRef]
- Zamorano, A.; Fiore, N. Draft Genome Sequence of 16SrIII-J Phytoplasma, a Plant Pathogenic Bacterium with a Broad Spectrum of Hosts. Genome Announc. 2016, 4, e01043-16. [Google Scholar] [CrossRef]
- Gamboa, C. Identificación de Efectores de Patogenicidad del Fitoplasma 16SrIII-J. Master’s Thesis, Universidad de Chile, Santiago, Chile, 2019; 43p. [Google Scholar]
- Zhang, H.; Bu, W. Exploring Large-Scale Patterns of Genetic Variation in the COI Gene among Insecta: Implications for DNA Barcoding and Threshold-Based Species Delimitation Studies. Insects 2022, 13, 703. [Google Scholar] [CrossRef]
- Serbina, L.Š.; Malenovský, I.; Queiroz, D.L.; Burckhardt, D. Jumping plant-lice of the tribe Paurocephalini (Hemiptera: Psylloidea: Liviidae) in Brazil. bioRxiv. 2024. [Google Scholar] [CrossRef]
- Wang, G.; Sun, C.; Hu, H.; Zhang, D.; Li, M. Complete Mitochondrial Genome of the Backswimmer: Notonecta triguttata Motschulsky, 1861 (Hemiptera: Notonectidae): Sequence, Structure, and Phylogenetic Analysis. Diversity 2024, 16, 16. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2006, 22, 148–155. [Google Scholar] [CrossRef]
- Sauvion, N.; Lachenaud, O.; Genson, G.; Rasplus, J.; Labonne, G. Are there several biotypes of Cacopsylla pruni? Bull. Insectol. 2007, 60, 185–186. [Google Scholar]
- Peccoud, J.; Labonne, G.; Sauvion, N. Molecular Test to Assign Individuals within the Cacopsylla pruni Complex. PLoS ONE 2013, 8, e76760. [Google Scholar] [CrossRef]
- Bodnár, D.; Koczor, S.; Tarcali, G.; Tóth, M.; Ott, P.G.; Tholt, G. Cacopsylla pruni (Hemiptera, Psyllidae) in an apricot orchard is more attracted to white sticky traps dependent on host phenology. Biodivers. Data J. 2022, 10, e82329. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The Plant Hypersensitive Response: Concepts, Control and Consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef]
- Wu, Z.; Qu, M.; Chen, M.; Lin, J. Proteomic and transcriptomic analyses of saliva and salivary glands from the Asian citrus psyllid, Diaphorina citri. J. Proteom. 2021, 238, 104137. [Google Scholar] [CrossRef]
- Weintraub, P.; Beanland, L. Insect Vectors of Phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Picciau, L.; Orrù, B.; Mandrioli, M.; Gonella, E.; Alma, A. Ability of Euscelidius variegatus to Transmit Flavescence Dorée Phytoplasma with a Short Latency Period. Insects 2020, 11, 603. [Google Scholar] [CrossRef]
- Jarausch, B.; Tedeschi, R.; Sauvion, N.; Gross, J.; Jarausch, W. Psyllid Vectors. In Phytoplasmas: Plant Pathogenic Bacteria—II: Transmission and Management of Phytoplasma-Associated Diseases; Bertaccini, A., Weintraub, P., Rao, G., Mori, N., Eds.; Springer: Cham, Switzerland, 2019; pp. 53–78. [Google Scholar]
- Trivellone, V.; Dietrich, C. Evolutionary Diversification in Insect Vector–Phytoplasma–Plant Associations. Ann. Entomol. Soc. Am. 2021, 114, 137–150. [Google Scholar] [CrossRef]
- Ku, C.; Lo, W.S.; Kuo, C.H. Horizontal Transfer of Potential Mobile Units in Phytoplasmas. Mob. Genet. Elem. 2013, 3, 4. [Google Scholar] [CrossRef]
- Dickinson, M. Mobile Units of DNA in Phytoplasma Genomes. Mol. Microbiol. 2010, 77, 1351–1353. [Google Scholar] [CrossRef]
- Fernández, F.; Guzmán, F.; Conci, L. Draft Genome Sequence of Cicuta Witches’ Broom Phytoplasma, Subgroup 16SrIII-J: A Subgroup with Phytopathological Relevance in South America. Trop. Plant Pathol. 2024, 49, 558–565. [Google Scholar] [CrossRef]
- Wei, W.; Inaba, J.; Zhao, Y.; Mowery, J.; Hammond, R. Phytoplasma Infection Blocks Starch Breakdown and Triggers Chloroplast Degradation, Leading to Premature Leaf Senescence, Sucrose Reallocation, and Spatiotemporal Redistribution of Phytohormones. Int. J. Mol. Sci. 2022, 23, 1810. [Google Scholar] [CrossRef]
- Ermacora, P.; Osler, R. Symptoms of Phytoplasma Diseases. Methods Mol. Biol. 2019, 1875, 53–67. [Google Scholar] [PubMed]
- Janik, K.; Mittelberger, C.; Moser, M. Lights Out: The Chloroplast Under Attack During Phytoplasma Infection? Annu. Plant Rev. 2024, 3, 399–426. [Google Scholar]
- Burckhardt, D. Jumping plant lice (Homoptera: Psylloidea) of the temperate neotropical region. Part 1: Psyllidae (subfamilies Aphalarinae, Rhinocolinae and Aphalaroidinae). Zool. J. Linn. Soc. 1987, 89, 299–392. [Google Scholar] [CrossRef]
- Burckhardt, D. Generic key to Chilean jumping plant-lice (Homoptera: Psylloidea) with includsion of potential exotic pests. Rev. Chil. Entomología 1994, 21, 57–67. [Google Scholar]
- Burckhardt, D. Psylloidea. In Biodiversidad de Artrópodos Argentinos; Roig-Juñet, S., Claps, L., Debandi, G., Eds.; SAE Ediciones: Hong Kong, China, 2008; Volume 2, pp. 189–199. [Google Scholar]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–479. [Google Scholar] [CrossRef]
- Barragán, G.; Almaraz, N.; Álvarez, R.; Delgado, E.; Pérez, J. DNA Isolation from Diabrotica virgifera zeae Krysan and Smith and Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) by a CTAB Simplified Procedure. Southwest. Entomol. 2009, 34, 289–294. [Google Scholar] [CrossRef]
- Schaff, D.; Lee, I.; Davis, R. Sensitive Detection and Identification of Mycoplasma-Like Organisms in Plants by Polymerase Chain Reactions. Biochem. Biophys. Res. Commun. 1992, 186, 1503–1509. [Google Scholar] [CrossRef]
- Gundersen, D.; Lee, I. Ultrasensitive Detection of Phytoplasmas by Nested-PCR Assays Using Two Universal Primer Pairs. Phytopathol. Mediterr. 1996, 35, 114–151. [Google Scholar]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA Genes from Culturable and Nonculturable Mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Kirkpatrick, B.C. Phytoplasma-Specific PCR Primers Based on Sequences of the 16S-23S rRNA Spacer Region. Appl. Environ. Microbiol. 1996, 62, 2988–2993. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. In The Mitochondrial Genome of Animals; MacIntyre, R.J., Ed.; Plenum Press: New York, NY, USA, 1994; pp. 95–130. [Google Scholar]
- Percy, D.M.; Crampton-Platt, A.; Sveinsson, S.; Lemmon, A.R.; Lemmon, E.M.; Ouvrard, D.; Burckhardt, D. Resolving the Psyllid Tree of Life: Phylogenomic Analyses of the Superfamily Psylloidea (Hemiptera). Syst. Entomol. 2018, 43, 762–776. [Google Scholar] [CrossRef]
- Sumner-Kalkun, J.C.; Sjölund, M.J.; Arnsdorf, Y.M.; Carnegie, F.; Highet, D.; Ouvrard, D.; Greenslade, A.F.C.; Bell, J.R.; Sigvald, R.; Kenyon, D.M. Insights into Phytoplasma Vector Ecology: A Global Overview. PLoS ONE 2020, 15, e0230741. [Google Scholar]
- Cho, G.; Malenovský, I.; Burckhardt, D.; Inoue, H.; Lee, S. DNA Barcoding of Pear Psyllids (Hemiptera: Psylloidea: Psyllidae), a Tale of Continued Misidentifications. Bull. Entomol. Res. 2020, 110, 521–534. [Google Scholar] [CrossRef]
- Šafářová, D.; Zrníková, E.; Holušová, K.; Ouředníčková, J.; Starý, M.; Navrátil, M. Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’. Agronomy 2023, 13, 2210. [Google Scholar] [CrossRef]
- Bastin, S.; Percy, D.M.; Siverio, F. Establishing reliable DNA barcoding primers for jumping plant lice (Psylloidea, Hemiptera). BMC Res. Notes 2023, 16, 322. [Google Scholar] [CrossRef]
- Wamonje, F.O.; Zhou, N.; Bamrah, R.; Wist, T.; Prager, S.M. Detection and Identification of a ‘Candidatus Liberibacter solanacearum’ Species from Ash Tree Infesting Psyllids. Phytopathology 2022, 112, 76–80. [Google Scholar] [CrossRef]
- Kuznetsova, V.G.; Labina, E.S.; Shapoval, N.A.; Maryańska-Nadachowska, A.; Lukhtanov, V.A. Cacopsylla fraudatrix sp. n. (Hemiptera: Psylloidea) Recognized from Testis Structure and Mitochondrial Gene COI. Entomol. Sci. 2012, 3547, 1–13. [Google Scholar]
- Gwiazdowski, R.A.; Foottit, R.G.; Maw, H.E.L.; Hebert, P.D.N. The Hemiptera (Insecta) of Canada: Constructing a Reference Library of DNA Barcodes. PLoS ONE 2015, 10, e0125635. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; He, Y.; Wang, X.; Gu, X. The Complete Mitochondrial Genome of Cyamophila willieti (Wu) (Hemiptera: Psyllidae). Mitochondrial DNA Part B 2019, 4, 3758–3759. [Google Scholar] [CrossRef]
- Jo, E.; Cho, G. The Complete Mitochondrial Genome of Cacopsylla burckhardti (Hemiptera, Psylloidea, Psyllidae). Biodivers. Data J. 2022, 10, e85094. [Google Scholar] [CrossRef] [PubMed]
- Que, S.; Yu, L.; Xin, T.; Zou, Z.; Hu, L.; Xia, B. Complete Mitochondrial Genome of Cacopsylla coccinae (Hemiptera: Psyllidae). Mitochondrial DNA Part B 2016, 27, 3169–3170. [Google Scholar] [CrossRef]
- Kang, A.R.; Kim, M.J.; Park, J.S.; Seo, H.-J.; Song, J.-H.; Won, K.-H.; Choi, E.D.; Kim, I. Comparative Analysis of Two Pear Pests, Cacopsylla jukyungi and Cacopsylla burckhardti (Hemiptera: Psyllidae), Based on Complete Mitochondrial Genomes and Comparison to Confamilial Species. Agronomy 2022, 12, 2037. [Google Scholar] [CrossRef]
- Wu, F.; Cen, Y.; Wallis, C.M.; Trumble, J.T.; Prager, S.; Yokomi, R.; Zheng, Z.; Deng, X.; Chen, J.; Liang, G. The Complete Mitochondrial Genome Sequence of Bactericera cockerelli and Comparison with Three Other Psylloidea Species. PLoS ONE 2016, 11, e0155318. [Google Scholar] [CrossRef]
- Wang, Y.; Halbert, S.; Mohamed, S.; Delatte, H.; Reynaud, B.; Beattie, G.A.C.; Holford, P.; Lu, J.; Cen, Y. Mitochondrial Genomes Reveal Diverse Lineages of Diaphorina citri Kuwayama (Hemiptera: Sternorrhyncha: Psyllidae) in Kenya and La Réunion. Diversity 2021, 23, 3109–3117. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, L.; Baumann, P. Organization of the Mitochondrial Genomes of Whiteflies, Aphids, and Psyllids (Hemiptera, Sternorrhyncha). BMC Evol. Biol. 2004, 4, 25. [Google Scholar] [CrossRef]
- Zhang, Q.L.; Guo, Z.L.; Yuan, M.L. The complete mitochondrial genome of Poratrioza sinica (Insecta: Hemiptera: Psyllidae). Mitochondrial DNA Part A 2014, 27, 734–735. [Google Scholar] [CrossRef]
- Wang, J.J.; Yang, M.F.; Dai, R.H.; Li, H.; Wang, X.Y. Characterization and Phylogenetic Implications of the Complete Mitochondrial Genome of Idiocerinae (Hemiptera: Cicadellidae). Int. J. Biol. Macromol. 2018, 120, 2366–2372. [Google Scholar] [CrossRef] [PubMed]


| Collection N° | Start Date of the Transmission Trial | Number of Immatures/Adults Collected | Number of Adults Used in Transmission Trials | TT Initiated |
|---|---|---|---|---|
| 1 | 4 October 2023 | 15/100 | 98 | 14 |
| 2 | 17 October 2023 | 0/70 | 70 | 9 |
| 3 | 3 November 2023 | 0/50 | 44 | 4 |
| 4 | 16 November 2023 | 0/60 | 60 | 6 |
| 5 | 30 November 2023 | 12/105 | 90 | 9 |
| 6 | 27 December 2023 | 10/110 | 99 | 9 |
| 7 | 24 January 2024 | 0/80 | 80 | 8 |
| 8 | 7 February 2024 | 0/94 | 70 | 7 |
| 9 | 21 February 2024 | 14/115 | 96 | 8 |
| 10 | 6 March 2024 | 100/600 | 96 | 8 |
| 11 | 18 March 2024 | 58/200 | 320 | 16 |
| 12 | 5 April 2024 | 66/160 | 160 | 8 |
| 13 | 16 April 2024 | 0/50 | 50 | 5 |
| 14 | 17 May 2024 | 0/20 | 20 | 2 |
| Total individuals | 275/1814 | 1353 | 113 | |
| Phytoplasma-Positive Plant Code at the End of TT | TT Start Date | Months After the Start of TT in Which the Plant Was Detected Positive for 16SrIII-J | Closest Phytoplasma with BLASTn | Sequence % Identity | Accession Number |
|---|---|---|---|---|---|
| TT 34 | 30 November 2023 | 3 | SBYWDP | 100 | PV053118 |
| TT 35 | 30 November 2023 | 6 | SBYWDP | 99.82 | PV053117 |
| TT 36 | 30 November 2023 | 6 | SBYWDP | 100 | PV053116 |
| TT 40 | 30 November 2023 | 7 | SBYWDP | 99.82 | PV053115 |
| TT 50 | 27 December 2023 | 7 | SBYWDP | 100 | PV053114 |
| TT 56 | 24 January 2024 | 7 | SBYWDP | 100 | PV053113 |
| TT 58 | 24 January 2024 | 7 | SBYWDP | 99.91 | PV053112 |
| Insect Code | Acc. Number | Closest species with BLASTn COI | % Coverage | % Identity | Acc. Number |
|---|---|---|---|---|---|
| ZTT 34 | PV055068 | Russelliana solanicola | 99 | 84.84 | PV055068 |
| ZTT 35 | PV055070 | Russelliana solanicola | 99 | 85.02 | PV055070 |
| ZTT 36 | PV055069 | Russelliana solanicola | 99 | 85.02 | PV055069 |
| ZTT 40 | PV055074 | Russelliana solanicola | 99 | 85.02 | PV055074 |
| ZTT 50 | PV055071 | Russelliana solanicola | 99 | 85.02 | PV055071 |
| ZTT 56 | PV055072 | Russelliana solanicola | 99 | 85.02 | PV055072 |
| ZTT 58 | PV055073 | Russelliana solanicola | 99 | 85.02 | PV055073 |
| Mallow Psyllid Gene | R. solanicola Gene | % Nucleotide Identity |
|---|---|---|
| Ins ZEIP gen 1 (ND 1) | NAD 1 | 80.74 |
| Ins ZEIP gen 2 (CYTB) | CYTB | 79.93 |
| Ins ZEIP gen 3 (NAD 6) | NAD 6 | 75.61 |
| Ins ZEIP gen 4 (NAD 4) Ins ZEIP gen 5 (NAD 4L) | NAD 4 NAD 4L | 78.80 84.38 |
| Ins ZEIP gen 6 (NAD 5) | NAD 5 | 78.23 |
| Ins ZEIP gen 7 (NAD 3) | NAD 3 | 78.16 |
| Ins ZEIP gen 8 (COX 3) | COX 3 | 81.71 |
| Ins ZEIP gen 9 (ATP 6) Ins ZEIP gen 10 (ATP 8) | ATP 6 ATP 8 | 78.67 74.16 |
| Ins ZEIP gen 11 (COX 2) | COX 2 | 83.13 |
| Ins ZEIP gen 12 (COX 1) | COX 1 | 84.67 |
| Ins ZEIP gen 13 (NAD 2) | NAD 2 | 76.83 |
| Gene | Position | RNA Strand Orientation | Length (bp) | Initiation Codon | Termination Codon | Anticodon | Intergenic Nucleotides |
|---|---|---|---|---|---|---|---|
| tRNAI | 1–63 | + | 63 | *nd | nd | GAT | 0 |
| tRNAQ | 61–127 | − | 67 | nd | nd | TTG | −2 |
| tRNAM | 127–190 | + | 63 | nd | nd | CAT | −1 |
| NAD2 | 191–1159 | + | 969 | ATA | TAA | nd | 0 |
| tRNAW | 1163–1224 | + | 62 | nd | nd | TCA | 3 |
| tRNAC | 1227–1291 | − | 65 | nd | nd | GCA | 2 |
| tRNAY | 1293–1356 | − | 64 | nd | nd | GTA | 1 |
| COX1 | 1372–2904 | + | 1533 | ATG | TAA | nd | 15 |
| tRNAL2 | 2905–2969 | + | 65 | nd | nd | TAA | 0 |
| COX2 | 2970–3627 | + | 657 | ATG | TAA | nd | 0 |
| tRNAK | 3634–3703 | + | 70 | nd | nd | CTT | 6 |
| tRNAD | 3704–3767 | + | 64 | nd | nd | GTC | 0 |
| ATP8 | 3768–3920 | + | 153 | ATT | TAA | nd | 0 |
| ATP6 | 3914–4588 | + | 675 | ATG | TAA | nd | −7 |
| COX3 | 4588–5370 | + | 783 | ATG | TAA | nd | −1 |
| tRNAG | 5371–5432 | + | 62 | nd | nd | TCC | 0 |
| NAD3 | 5433–5783 | + | 351 | ATT | TAA | nd | 0 |
| tRNAA | 5787–5848 | + | 62 | nd | nd | TGC | 3 |
| tRNAR | 5852–5913 | + | 62 | nd | nd | TCG | 3 |
| tRNAN | 5913–5976 | + | 64 | nd | nd | GTT | −1 |
| tRNAS1 | 5977–6029 | + | 73 | nd | nd | GCT | 0 |
| tRNAE | 6030–6090 | + | 61 | nd | nd | TTC | 0 |
| tRNAF | 6079–6141 | − | 63 | nd | nd | GAA | −12 |
| NAD5 | 6142–7762 | − | 1620 | TTG | TAA | nd | 0 |
| tRNAH | 7763–7825 | − | 63 | nd | nd | GTG | 0 |
| NAD4 | 7821–9068 | − | 1248 | ATG | TAA | nd | −5 |
| NAD4L | 9062–9349 | − | 288 | TTG | TAG | nd | −7 |
| tRNAT | 9351–9412 | + | 62 | nd | nd | TGT | 1 |
| tRNAP | 9413–9474 | − | 62 | nd | nd | TGG | 0 |
| NAD6 | 9478–9963 | + | 486 | ATA | TAA | nd | 3 |
| CYTB | 9963–11,099 | + | 1136 | ATG | TAA | nd | −1 |
| tRNAS2 | 11,099–11,161 | + | 63 | nd | nd | TGA | −1 |
| NAD1 | 11,187–12,101 | − | 915 | ATA | TAA | nd | 26 |
| tRNAL1 | 12,102–12,166 | − | 65 | nd | nd | TAG | 0 |
| rRNAL | 12,172–13,318 | − | 1146 | nd | nd | nd | 5 |
| tRNAV | 13,315–13,375 | − | 61 | nd | nd | TAC | −3 |
| rRNAS | 13,374–14,119 | − | 746 | nd | nd | nd | −1 |
| Control Region | 14,120–14,835 | 735 | nd | nd | nd | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llantén, T.; Cabrera, S.; Fuentes, J.; Gamboa, C.; González, C.; Zamorano, A.; Curkovic, T.; Burckhardt, D.; Fiore, N. First Report of a Psyllid Vector of ‘Candidatus Phytoplasma pruni’ (Strain 16SrIII-J). Plants 2025, 14, 1279. https://doi.org/10.3390/plants14091279
Llantén T, Cabrera S, Fuentes J, Gamboa C, González C, Zamorano A, Curkovic T, Burckhardt D, Fiore N. First Report of a Psyllid Vector of ‘Candidatus Phytoplasma pruni’ (Strain 16SrIII-J). Plants. 2025; 14(9):1279. https://doi.org/10.3390/plants14091279
Chicago/Turabian StyleLlantén, Tomás, Sebastián Cabrera, Javiera Fuentes, Camila Gamboa, Constanza González, Alan Zamorano, Tomislav Curkovic, Daniel Burckhardt, and Nicola Fiore. 2025. "First Report of a Psyllid Vector of ‘Candidatus Phytoplasma pruni’ (Strain 16SrIII-J)" Plants 14, no. 9: 1279. https://doi.org/10.3390/plants14091279
APA StyleLlantén, T., Cabrera, S., Fuentes, J., Gamboa, C., González, C., Zamorano, A., Curkovic, T., Burckhardt, D., & Fiore, N. (2025). First Report of a Psyllid Vector of ‘Candidatus Phytoplasma pruni’ (Strain 16SrIII-J). Plants, 14(9), 1279. https://doi.org/10.3390/plants14091279







