Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled
Abstract
:1. Introduction
2. Results
2.1. Selection of Enhancer-Trap Lines
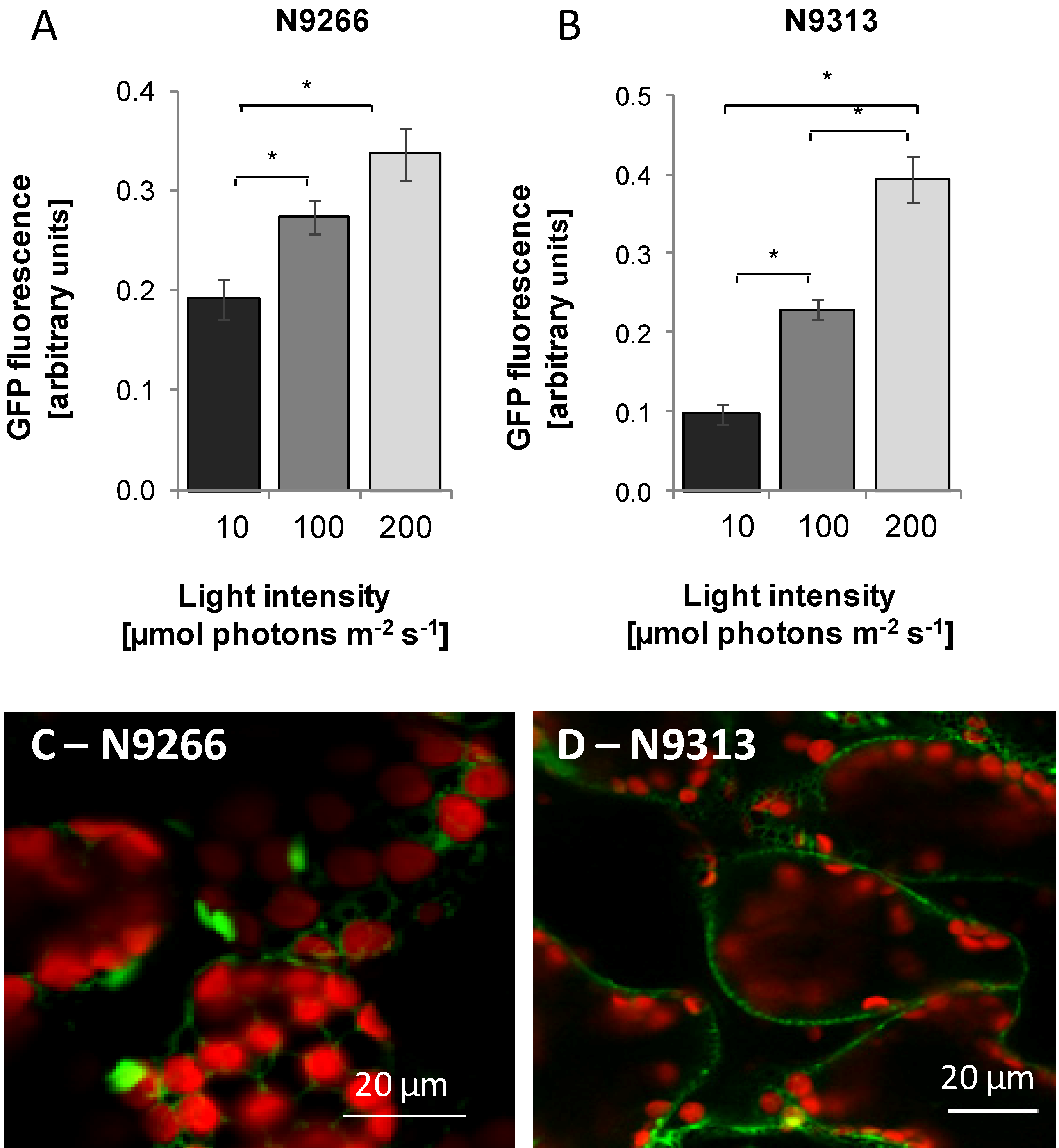
2.2. Identification of Enhancer Trapped Sequences and T-DNA Numbers

2.3. Light Quality Regulation of SIG5 by the Distal and Proximal Upstream Regions

2.4. Arabidopsis SIG5 Transcription Is Red Light Sensitive
2.5. In Silico Analysis of the SIG5 Promoter for Putative Light Responsive Elements

| Motif | Sequence a | Position b |
|---|---|---|
| AE-box | AGAAACAT (+) AGAAACAT (+) | −60 −1021 |
| GATA motifs | GATA (+) GATA (−) GATA (+) GATA (+)c GATA (−)c GATA (+) GATA (−) GATA (−) d GATA (−) d GATA (−) d GATA (+) d | −71 −163 −441 −841 −871 −914 −989 −1593 −1751 −1944 −1985 |
| Box 4 | ATTAAT (−) ATTAAT (−) ATTAAT (−) ATTAAT (−) | −593 −654 −962 −1428 |
| GA motif | AAGGAAGA (−) c | −707 |
| I-box | CACTTATGCT (−) c aAGATAAGA (−) | −732 −1587 |
| ACE | AAAACGTTTA (−) c CTAACGTATT (−) d ACGTGGA (−) d | −763 −1442 −1795 |
| Box I | TTTCAAA (−) TTTCAAA (−) TTTCAAA (+) | −917 −1751 −1901 |
| 3-AF1 binding site | AAGAGATATTT (−) | −922 |
| ATCT-motif | AATGTAATCT (+) AATGTAATCT (+) AATGTAATCT (+) | −1205 −1455 −1460 |
| L-box | AAATTAACCAAC (−) | −1426 |
| TCT-motif | TCTTAC (+) | −1541 |
| CATT-motif | GCATTC (+) | −1687 |
| GAG-motif | GGAGATG (−) | −1699 |
| GC-box | CACGTC (+)d | −1719 |
| G-box | CACGTG (−)d | −1794 |
2.6. Mapping Revealed Locally Restricted Blue-Light Sensitivity and Disperse Red-Light Sensitivity

2.7. Blue- and Red-Light Regulation of the Proximal Upstream Region
2.8. Photoreceptor Control of the Distal SIG5 Upstream Region


2.9. Regulation of the Distal SIG5 Upstream Region by HY5 and COP1

| cis-element | Sequence a | Position b | Introduced mutation c | Reference |
|---|---|---|---|---|
| G-box | CACGTG (+) | −1794 |  | [45] |
| GC-box | GACGTG (+) | −1719 |  | [45] |
| GATA-motif | GATAAG (+) | −1593 |  | [46] |
2.10. Yeast-1-Hybrid Analysis of HY5 Binding to the SIG5 Promoter
2.11. Impact of Photosynthetic Electron Transport on Light Regulation


2.12. Sugar Effect on SIG5 Transcription

3. Experimental
3.1. Plant Material and Growth Conditions
3.2. Extraction of Genomic DNA, TAIL-PCR and Sequence Analysis
3.3. Southern Blot Analysis
3.4. Identification and Isolation of Homozygous T-DNA Insertion Lines by PCR
| PCR reaction/ product | TA [°C] | Forward primer/reverse primer (5'→3') |
|---|---|---|
| TAIL-PCR, T-DNA primers | 73.1 | CACTTGGCGCACTTCGGCTTC |
| 67.5 | AGCTTCTTGAGGCGGCAGA | |
| 63.3 | GGAGCTTCATTGTTGGATCC | |
| TAIL-PCR, AD primers | 46.0 | NTCGA(G/C)T(A/T)T(G/C)G(A/T)GTT |
| 46.8 | NGTCGA(G/C)(A/T)GANA(A/T)GAA | |
| 34.8 | (A/T)GTGNAG(A/T)ANCANAGA | |
| Verification of N9266 T-DNA | 58 | CGTATCACGCGGCGC |
| Verification of N9313 T-DNA | 54 | CTCCGACTCTTGCGATAT |
| qRT-PCR, ACT2 | 60 | TCTTCCGCTCTTTCTTTCCAAGC |
| ACCATTGTCACACACGATTGGTTG | ||
| qRT-PCR, SIG5 | 60 | TGGAGCTAATAACAGCAGACAGC |
| TCGGCTTCAATGAATCGAGCAC | ||
| qRT-PCR, GFP | 60 | CCATTACCTGTCCACACAATC |
| GTTCATCCATGCCATGTG | ||
| qRT-PCR, HY5 | 60 | AGAACAAGCGGCTGAAGAGGTTG |
| TCCTCTCTCTTGCTTGCTGAGCTG | ||
| Construction of pSIG5wildtype | 45 | ACTAGTTTTTTCTGCAGGTAACTCCGACTCTTGCG |
| GCTTGAGAGATTACATTATT | ||
| Construction of Δ35S | 62 | TTCGCAAGACCCTTCCTCTATATAAGG |
| GGGTACCGGTCGCCACC | ||
| Analysis of SALK_015625 | 50 | CAATCATGGTTTAATTCGT a |
| GATCCACAACCACAAGCC | ||
| Analysis of SALK_077048 | 45 | GTTATTGATCTGTACCTAGC a |
| and 072457 | AAATACGATAGATGTGTTG | |
| Analysis of SALK_019261 | 45 | ATCACAATCTTAAGGCTCAAAA |
| AAATACGATAGATGTGTTG a | ||
| Analysis of SALK_133729 | 45 | ATCACAATCTTAAGGCTCAAAA a |
| AAATACGATAGATGTGTTG | ||
| Identification of cry2-1 allele | 58 | CAGTTTTATCCTGGAAGAGCCTC |
| CTTCTCCTTTACGGTATGGTCC | ||
| Identification of hy1-1 allele | 56 | GGAATTAGCAGAGAAGGATCC |
| TATCCGCTCTGCCACCTG | ||
| Identification of phyA-201 allele | 55 | CCTTAAATGAAGTGTTGACTGC |
| GCAAGATGCACAGAACG | ||
| Identification of phyB-5 allele | 55 | GTTGTGGAGTGGTTGCTTG |
| CATAGCCGCCTCAGATTC | ||
| Identification of phot1-5 allele | 58 | CCACTTGCAACCTATGCG |
| CTCTTTCACTGCGGTTTCTTC | ||
| Identification of phot2-1 allele | 54 | CTCTGCCTCACAATAAGGAG |
| CTGCCAGTATCACCAGAGC | ||
| Identification of SALK_056405C | 58 | GCGGTAGCCAGAGTAATCTATTCC |
| TCCTCTCTCTTGCTTGCTGAGCTG | ||
| ATTTTGCCGATTTCGGAAC b | ||
| Construction of pSIG5ΔGATA-1a | 60 | GGTGGCGACCGGTACC |
| CATCTTTTACTGAATACTTTGAGTTATTTGCACATATAG | ||
| Construction of pSIG5ΔGATA-1b | 55 | CTATATGTGCAAATAACTCAAAGTATTCAGTAAAAGATG |
| GTAACTCCGACTCTTGCGAT | ||
| Construction of pSIG5ΔG-1a | 60 | GGTGGCGACCGGTACC |
| CTGAGAAGACCATCCAATTGTATAATTCCTGATC | ||
| Construction of pSIG5ΔG-1b | 55 | GATCAGGAATTATACAATTGGATGGTCTTCTCAG |
| GTAACTCCGACTCTTGCGAT | ||
| Construction of pSIG5ΔGC-1a | 60 | GGTGGCGACCGGTACC |
| CTATAAATTGGCCAATTCGTCTCTCTCTC | ||
| Construction of pSIG5ΔGC-1b | 55 | GAGAGAGAGACGAATTGGCCAATTTATAG |
| GTAACTCCGACTCTTGCGAT | ||
| Construction of pSIG5 fragments-2 | 63 | GGTGGCGACCGGTACC |
| GTAACTCCGACTCTTGCGAT | ||
| Y1H-construction of pRBCS1A | 55 | TTTTTGAGCTCGATTTTGAGTGTGGATATGTGT |
| TTTTTGAATTCCCAGGCAAGTAAAATGAGCAAG | ||
| Y1H-construction of HY5 CDS | TTTTTGGATCCTACAGGAACAAGCGACTAGCTC c | |
| TTTTTCTCGAGTCAAAGGCTTGCATCAGC | ||
| Y1H-construction of pSIG5distal | 57 | TTTTTGAGCTCCACAATCTTAAGGCTCAAAAATTG |
| TTTTTGGGCCCTCGGATGCTTTACATGGTG | ||
| Y1H-construction of pSIG5proximal | 60 | TTTTTGAATTCGTAACTCCGACTCTTGCG |
| TTTTTGAGCTCGCTTGAGAGATTACATTATT |
3.5. Determination of Reporter Gene Activity
3.6. Transcript Abundance Analysis
3.7. mRNA Decay Analysis
3.8. Reporter Gene Construct Design, Site-Directed Mutagenesis and Tobacco Transfection
3.9. In Silico Analysis of Promoter Sequences
3.10. Yeast One-Hybrid Analysis
3.11. Crossing Arabidopsis thaliana Plants and Mutant Selection
3.12. Fluorescence Microscopy
3.13. Chlorophyll-a Fluorescence Analysis
4. Discussion
4.1. Transcriptional Regulation of SIG5 Promoter
4.2. Blue-light Regulation
4.3. Red-light Sensitivity
4.4. Antero- and Retrograde Signaling Connecting Chloroplast and Nuclear Gene Expression
5. Conclusions

Acknowledgements
Abbreviations
| 3-AT | 3-amino-1,2,4-triazol |
| Act D | actinomycin D |
| ANOVA | analysis of variance |
| BL | blue light |
| BLRP | blue light responsive promoter |
| CDS | coding sequence |
| DCMU | 3-(3,4-Dichlorophenyl)-1,1-dimethylurea |
| ET | enhancer trap FR far red |
| FRL | far-red light |
| GFP | green fluorescent protein |
| MS | Murashige and Skoog medium |
| PCR | polymerase chain reaction |
| PSII | photosystem II |
| qRT-PCR | quantitative real-time PCR |
| RL | red light |
| RUBISCO | ribulose-1,5-bisphosphate carboxylase/oxygenease |
| SD | synthetic dropout |
| SIG5 | sigma factor 5 |
| TAIL-PCR | thermal asymmetric interlaced PCR |
| UAS | upstream activation sequence |
| UTR | untranslated region |
| Y1H | yeast-one-hybrid |
Author Contributions
Conflicts of Interest
References
- Terzaghi, W.B.; Cashmore, A.R. Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 445–474. [Google Scholar]
- Troxler, R.F.; Zhang, F.; Hu, J.; Bogorad, L. Evidence that sigma factors are components of chloroplast RNA polymerase. Plant Physiol. 1994, 104, 753–759. [Google Scholar]
- Block, A.; Dangl, J.L.; Hahlbrock, K.; Schulze-Lefert, P. Functional borders, genetic fine-structure, and distance requirements of cis elements mediating light responsiveness of the parsley chalcone cynthase promoter. Proc. Natl. Acad. Sci. USA 1990, 87, 5387–5391. [Google Scholar] [CrossRef]
- Gilmartin, P.M.; Sarokin, L.; Memelink, J.; Chua, N.-H. Molecular light switches for plant genes. Plant Cell 1990, 2, 369–378. [Google Scholar]
- Giuliano, G.; Pichersky, E.; Malik, V.S.; Timko, M.P.; Scolnik, P.A.; Cashmore, A.R. An evolutionarily conserved protein-binding sequence upstream of a plant light-regulated gene. Proc. Natl Acad. Sci. USA 1988, 85, 7089–7093. [Google Scholar]
- Ha, S.B.; An, G.H. Identification of upstream regulatory elements involved in the developmental expression of the Arabidopsis thaliana cab1 gene. Proc. Natl. Acad. Sci. USA 1988, 85, 8017–8021. [Google Scholar] [CrossRef]
- Borello, U.; Ceccarelli, E.; Giuliano, G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different L-box elements in plant light-regulated promoters. Plant J. 1993, 4, 611–619. [Google Scholar]
- Grob, U.; Stuber, K. Discrimination of phytochrome dependent light inducible from non-light inducible plant genes—Prediction of a common light-responsive element (Lre) in phytochrome dependent light inducible plant genes. Nucleic Acids Res. 1987, 15, 9957–9973. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, M.; Kumar, V.; Pandey, B.; Singh, S.P.; Bag, S.K.; Singh, B.D.; Tuli, R.; Sawant, S.V. Distinct role of core promoter architecture in regulation of light-mediated responses in plant genes. Mol. Plant 2014, 7, 626–641. [Google Scholar] [CrossRef]
- Martinez-Garcia, J.F.; Huq, E.; Quail, P.H. Direct targeting of light signals to a promoter element-bound transcription factor. Science 2000, 288, 859–863. [Google Scholar] [CrossRef]
- Rüdiger, W.; Oster, U. Intracellular signaling from plastids to the nucleus. In Photosynthesis, 34th ed.; Eaton-Rye, J.J., Tripathy, B.C., Sharkey, T.D., Eds.; Springer: Dortrecht, The Netherlands, 2012; pp. 175–190. [Google Scholar]
- Chory, J. Light signal transduction: An infinite spectrum of possibilities. Plant J. 2010, 61, 982–991. [Google Scholar] [CrossRef]
- Nelson, T.; Harpster, M.H.; Mayfield, S.P.; Taylor, W.C. Light-regulated gene expression during maize leaf development. J. Cell Biol. 1984, 98, 558–564. [Google Scholar] [CrossRef]
- Lorrain, S.; Genould, T.; Fankhauser, C. Let there be light in the nucleus! Curr. Opin. Plant Biol. 2006, 9, 509–514. [Google Scholar] [CrossRef]
- Millar, A.J. Input signals to the plant circadian clock. J. Exp. Bot. 2004, 55, 277–283. [Google Scholar] [CrossRef]
- Sullivan, J.A.; Deng, X.W. From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 2003, 260, 289–297. [Google Scholar] [CrossRef]
- Larkin, R.M.; Ruckle, M.E. Integration of light and plastid signals. Curr. Opin. Plant Biol. 2008, 11, 593–599. [Google Scholar] [CrossRef]
- Neff, M.M.; Chory, J. Genetic Interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998, 118, 27–35. [Google Scholar] [CrossRef]
- Chen, F.; Li, B.; Li, G.; Charron, J.B.; Dai, M.; Shi, X.; Deng, X.W. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 2014, 26, 1949–1966. [Google Scholar] [CrossRef]
- Haseloff, J. GFP variants for multispectral imaging of living cells. Methods Cell Biol. 1999, 58, 139–151. [Google Scholar] [CrossRef]
- Laplaze, L.; Parizot, B.; Baker, A.; Ricaud, L.; Martiniére, A.; Auguy, F.; Franche, C.; Nussaume, L.; Bogusz, D.; Haseloff, J. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 2433–2442. [Google Scholar] [CrossRef]
- He, Y.H.; Tang, W.N.; Swain, J.D.; Green, A.L.; Jack, T.P.; Gan, S.S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001, 126, 707–716. [Google Scholar] [CrossRef]
- Gardner, M.J.; Baker, A.J.; Assie, J.M.; Poethig, R.S.; Haseloff, J.P.; Webb, A.A.R. GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J. Exp. Bot. 2009, 60, 213–226. [Google Scholar]
- Tsunoyama, Y.; Ishizaki, Y.; Morikawa, K.; Kobori, M.; Nakahira, Y.; Takeba, G.; Toyoshima, Y.; Shiina, T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSIG5. Proc. Natl. Acad. Sci. USA 2004, 101, 3304–3309. [Google Scholar] [CrossRef]
- Noordally, Z.B.; Ishii, K.; Atkins, K.A.; Wetherill, S.J.; Kusakina, J.; Walton, E.J.; Kato, M.; Azuma, M.; Tanaka, K.; Hanaoka, M.; et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 2013, 339, 1316–1319. [Google Scholar] [CrossRef]
- Pena-Ahumada, A.; Kahmann, U.; Dietz, K.J.; Baier, M. Regulation of peroxiredoxin expression versus expression of Halliwell-Asada-Cycle enzymes during early seedling development of Arabidopsis thaliana. Photosyn. Res. 2006, 89, 99–112. [Google Scholar] [CrossRef]
- Liu, Y.G.; Mitsukawa, N.; Oosumi, T.; Whittier, R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995, 8, 457–463. [Google Scholar]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic fluorescence pistograph” browser for exploring and analyzing large-scale data sets. PLoS One 2007, 2, e178. [Google Scholar] [CrossRef]
- Mochizuki, T.; Onda, Y.; Fujiwara, E.; Wada, M.; Toyoshima, Y. Two independent light signals cooperate in the activation of the plastid psbD blue light-responsive promoter in Arabidopsis. FEBS Lett. 2004, 571, 26–30. [Google Scholar] [CrossRef]
- Onda, Y.; Yagi, Y.; Saito, Y.; Takenaka, N.; Toyoshima, Y. Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: Differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J. 2008, 55, 968–978. [Google Scholar] [CrossRef]
- Tsunoyama, Y.; Morikawa, K.; Shiina, T.; Toyoshima, Y. Blue light specific and differential expression of a plastid sigma factor, SIG5 in Arabidopsis thaliana. FEBS Lett. 2002, 516, 225–228. [Google Scholar] [CrossRef]
- Nagashima, A.; Hanaoka, M.; Shikanai, T.; Fujiwara, M.; Kanamaru, K.; Takahashi, H.; Tanaka, K. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 357–368. [Google Scholar]
- Oh, S.; Montgomery, B.L. Phytochrome-dependent coordinate control of distinct aspects of nuclear and plastid gene expression during anterograde signaling and photomorphogenesis. Front. Plant Sci. 2014, 5, e171. [Google Scholar]
- Rombauts, S.; Déhais, P.; van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.M.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Kagawa, T.; Kasahara, M.; Abe, T.; Yoshida, S.; Wada, M. Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol. 2004, 45, 416–426. [Google Scholar]
- Guo, H.; Duong, H.; Ma, N.; Lin, C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 1999, 19, 279–287. [Google Scholar] [CrossRef]
- Lopez-Juez, E.; Dillon, E.; Magyar, Z.; Khan, S.; Hazeldine, S.; de Jager, S.M.; Murray, J.A.H.; Beemster, G.T.S.; Bogre, L.; Shanahan, H. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 2008, 20, 947–968. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, E.; Haupt, W. Blue light effects in phytocrome-mediated responses. In Encyclopedia of Plant Physiology; Shropshire, W., Jr., Mors, H., Eds.; Springer: New York, NY, USA, 1983; p. 16. [Google Scholar]
- Muramoto, T.; Kohchi, T.; Yokota, A.; Hwang, I.H.; Goodman, H.M. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 1999, 11, 335–347. [Google Scholar] [CrossRef]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Puente, P.; Deng, X.W.; Wei, N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998, 15, 69–77. [Google Scholar] [CrossRef]
- Montemartini, M.; Kalisz, H.M.; Hecht, H.J.; Steinert, P.; Flohe, L. Activation of active-site cysteine residues in the peroxiredoxin-type tryparedoxin peroxidase of Crithidia fasciculata. Eur. J. Biochem. 1999, 264, 516–524. [Google Scholar] [CrossRef]
- McKendree, W.L., Jr.; Ferl, R.J. Functional elements of the Arabidopsis Adh promoter include the G-box. Plant Mol. Biol. 1992, 19, 859–862. [Google Scholar] [CrossRef]
- Donald, R.G.; Cashmore, A.R. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990, 9, 1717–1726. [Google Scholar]
- Laisk, A.; Oja, V.; Eichelmann, H.; Dall’Osto, L. Action spectra of photosystems II and I and quantum yield of photosynthesis in leaves in State 1. Biochim. Biophys. Acta 2014, 1837, 315–325. [Google Scholar]
- Pfannschmidt, T.; Nilsson, A.; Allen, J.F. Photosysnthetic control of chloroplast gene expression. Nature 1999, 397, 625–628. [Google Scholar] [CrossRef]
- Trebst, A.; Depka, B.; Kraft, B.; Johanningmeier, U. The QB site modulates the conformation of the photosystem II reaction center polypeptides. Photosyn. Res. 1988, 18, 163–177. [Google Scholar]
- Rook, F.; Corke, F.; Baier, M.; Holman, R.; May, A.G.; Bevan, M.W. Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J. 2006, 46, 1045–1058. [Google Scholar] [CrossRef]
- NCBI-Server. Available online: http://www.ncbi.nlm.nih.gov (accessed on 18 April 2010).
- Sheffield, J.B. ImageJ, a useful tool for biological image processing and analysis. Microsc. Microanal. 2007, 13, 200–201. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll-a and chlorophyll-b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Baier, M.; Ströher, E.; Dietz, K.J. The acceptor availability at photosystem I and ABA control nuclear expression of 2-Cys peroxiredoxin-A in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 997–1006. [Google Scholar]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, e465. [Google Scholar] [CrossRef]
- Quantprime. Available online: http://www.quantprime.mpimp-golm.mpg.de (accessed on 28 January 2010).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- An, Y.Q.; McDowell, J.M.; Huang, S.; McKinney, E.C.; Chambliss, S.; Meagher, R.B. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996, 10, 107–121. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Ross, J. Messenger-RNA stability in mammalian cells. Microbiol. Rev. 1995, 59, 423–450. [Google Scholar]
- Gutierrez, R.A.; Ewing, R.M.; Cherry, J.M.; Green, P.J. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 2002, 99, 11513–11518. [Google Scholar] [CrossRef]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. TIPS 2002, 7, 193–195. [Google Scholar]
- Koncz, C.; Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Arabidopsis: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2002. [Google Scholar]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef]
- English, J.J.; Davenport, G.F.; Elmayan, T.; Vaucheret, H.; Baulcombe, D.C. Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 1997, 12, 597–603. [Google Scholar] [CrossRef]
- PLACE. Available online: http://dna.affrc.go.jp/PLACE/signal-scan.html (accessed on 8 May 2011).
- PlantCARE. Available online: http://bioinformatics.psb.ugent.be/web-tools/plantcare/html (accessed on 8 May 2011).
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Lascéve, G.; Leymarie, J.; Olney, M.A.; Liscum, E.; Christie, J.M.; Vavasseur, A.; Briggs, W.R. Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol. 1999, 120, 605–614. [Google Scholar] [CrossRef]
- LSM Image Browser Software, Carl Zeiss: Jena, Germany, 2009.
- Schreiber, U.; Bilger, W. Progress in chlorophyll fluorescence research: Major developments during the past years in retrospect. Prog. Bot. 1993, 54, 151–173. [Google Scholar]
- Tanaka, K.; Tozawa, Y.; Mochizuki, N.; Shinozaki, K.; Nagatani, A.; Wakasa, K.; Takahashi, H. Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: Evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 1997, 413, 309–313. [Google Scholar] [CrossRef]
- Fujiwara, M.; Nagashima, A.; Kanamaru, K.; Tanaka, K.; Takahashi, H. Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase [sigma] factors in Arabidopsis thaliana. FEBS Lett. 2000, 481, 47–52. [Google Scholar] [CrossRef]
- Sugiura, M. The Chloroplast Chromosomes in Land Plants. Annu. Rev. Cell. Biol. 1989, 5, 51–70. [Google Scholar] [CrossRef]
- Hoffer, P.H.; Christopher, D.A. Structure and blue-light-responsive transcription of a chloroplast psbD promoter from Arabidopsis thaliana. Plant Physiol. 1997, 115, 213–222. [Google Scholar]
- Thum, K.E.; Kim, M.; Morishige, D.T.; Eibl, C.; Koop, H.U.; Mullet, J.E. Analysis of barley chloroplast psbD light-responsive promoter elements in transplastomic tobacco. Plant Mol. Biol. 2001, 47, 353–366. [Google Scholar] [CrossRef]
- Kanamaru, K.; Nagashima, A.; Fujiwara, M.; Shimada, H.; Shirano, Y.; Nakabayashi, K.; Shibata, D.; Tanaka, K.; Takahashi, H. An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol. 2001, 42, 1034–1043. [Google Scholar]
- Ishizaki, Y.; Ozono, K.; Takenaka, C.; Tsunoyama, Y.; Nakahira, Y.; Tanaka, K.; Kanamaru, K.; Hanaoka, M.; Shiina, T. An Arabidopsis double mutant sig2sig6 exhibits albino phenotype. Plant Cell Physiol. 2006, 47, S187. [Google Scholar]
- Shalitin, D.; Yu, X.; Maymon, M.; Mockler, T.; Lin, C. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 2003, 15, 2421–2429. [Google Scholar] [CrossRef]
- Shalitin, D.; Yang, H.; Mockler, T.C.; Maymon, M.; Guo, H.; Whitelam, G.C.; Lin, C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 2002, 417, 763–767. [Google Scholar] [CrossRef]
- Sharrock, R.A.; Clack, T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002, 130, 442–456. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Pfannschmidt, T.; Nilsson, A.; Tullberg, A.; Link, G.; Allen, J.F. Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life 1999, 48, 271–276. [Google Scholar] [CrossRef]
- Tullberg, A.; Alexciev, K.; Pfannschmidt, T.; Allen, J.F. Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 2000, 41, 1045–1054. [Google Scholar]
- Koch, K.E. Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 47, 509–540. [Google Scholar]
- Allen, J.F.; Pfannschmidt, T. Balancing the two photosystems: Photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B 2000, 355, 1351–1357. [Google Scholar] [CrossRef]
- Tikkanen, M.; Gollan, P.J.; Suorsa, M.; Kangasjarvi, S.; Aro, E.M. STN7 operates in retrograde signaling through controlling redox balance in the electron transfer chain. Front. Plant Sci. 2012, 3, e277. [Google Scholar]
- Aro, E.M.; Virgin, I.; Andersson, B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1993, 1143, 113–134. [Google Scholar] [CrossRef]
- Keren, N.; Berg, A.; van Kan, P.J.; Levanon, H.; Ohad, I. Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: The role of back electron flow. Proc. Natl. Acad. Sci. USA 1997, 94, 1579–1584. [Google Scholar]
Appendix


© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mellenthin, M.; Ellersiek, U.; Börger, A.; Baier, M. Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled. Plants 2014, 3, 359-391. https://doi.org/10.3390/plants3030359
Mellenthin M, Ellersiek U, Börger A, Baier M. Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled. Plants. 2014; 3(3):359-391. https://doi.org/10.3390/plants3030359
Chicago/Turabian StyleMellenthin, Marina, Ulrike Ellersiek, Anna Börger, and Margarete Baier. 2014. "Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled" Plants 3, no. 3: 359-391. https://doi.org/10.3390/plants3030359
APA StyleMellenthin, M., Ellersiek, U., Börger, A., & Baier, M. (2014). Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled. Plants, 3(3), 359-391. https://doi.org/10.3390/plants3030359




