Abstract
Plant immunity relies on a complex network of hormone signaling pathways in which jasmonic acid (JA) plays a central role. Successful microbial pathogens or symbionts have developed strategies to manipulate plant hormone signaling pathways to cause hormonal imbalances for their own benefit. These strategies include the production of plant hormones, phytohormone mimics, or effector proteins that target host components to disrupt hormonal signaling pathways and enhance virulence. Here, we describe the molecular details of the most recent and best-characterized examples of specific JA hormonal manipulation by microbes, which exemplify the ingenious ways by which pathogens can take control over the plant’s hormone signaling network to suppress host immunity.
1. Introduction
Agricultural productivity depends on the capacity of plants to adapt to changing environmental conditions. In nature, plants live in complex environments in which they intimately interact with a broad range of microbial pathogens with different infection strategies. To defend themselves, plants have evolved sophisticated strategies to perceive their attacker during the infection process and to translate this perception into an effective immune response. Recognition of highly conserved microbe-associated molecular patterns (MAMPs) by host cell transmembrane pattern recognition receptors (PRRs) leads to MAMP-triggered immunity (MTI) that restricts pathogen growth [1]. To overcome such lines of defense, adapted pathogens deliver phytotoxins and virulence molecules called effector proteins into the plant cell to promote pathogenesis in a process known as effector-triggered susceptibility (ETS) [1]. Inside the plant cell, phytotoxins and effectors target host molecules to subvert the host cell physiology and disrupt defenses. However, despite the fact that elucidating effector action is essential to understanding bacterial pathogenesis, the molecular function and host targets of the vast majority of effectors remain largely unknown.
Plant immunity relies on a complex network of hormone signaling pathways in which jasmonic acid (JA) and salicylic acid (SA) play a central role [2,3]. Both molecules control defense responses to different types of microbes and thus, they orchestrate a different and complex transcriptional reprogramming response that eventually leads to plant resistance. The JA pathway is primarily induced by and effective in mediating resistance against herbivores and necrotrophic pathogens such as Botrytis cinerea, whereas the SA pathway is primarily induced by and effective in mediating resistance against biotrophic and hemi-biotrophic pathogens such as Pseudomonas syringae [3]. JA and SA defense pathways generally antagonize each other and thus, elevated resistance against necrotrophs is often correlated with increased susceptibility to biotrophs and vice versa [4]. JAs play a key role in plant survival by contributing to adaptation to biotic and abiotic stresses and modulating many physiological and developmental agricultural traits such as root growth and fertility [5]. In the complex picture of hormonal plant immunity, other hormones such as abscisic acid (ABA), auxins, brassinosteroids (BRs), cytokinins (CKs), ethylene (ET), gibberellic acid (GAs), and oxylipins (other than JA) function as modulators of the plant immune signaling network as well, fine-tuning the hormonal balances to optimize resistance to the invading organism [2,6,7]. Therefore, the collective contribution of hormones during plant-pathogen interactions is crucial to establish the robust immune transcriptional program against the specific infection strategy of invading attackers.
In the last decade, a combination of genetic, molecular, and biochemical analyses have dissected the core JA signal transduction components linking JA synthesis to hormone-induced changes in gene expression. Among all JAs found in nature, (+)-7-iso-JA-l-Ile (JA-Ile) is the molecularly active form of the JA hormone [8]. JA-Ile is perceived through a co-receptor complex formed by the F-box protein CORONATINE-INSENSITIVE 1 (COI1) and JASMONATE ZIM DOMAIN (JAZ) proteins [9,10,11,12]. COI1 controls the turnover of the JAZ co-receptors in response to JA-Ile. The JAZ family is composed of 12 canonical members in Arabidopsis and an atypical repressor, JAZ13 [13,14]. JAZ proteins are repressors of the JA-signaling transcription factors (TFs), such as the basic helix-loop-helix (bHLH) MYC2, MYC3 and MYC4, that control JA-regulated genes [9,10,11,15,16,17]. To initiate transcription, MYCs physically interacts with the MEDIATOR25 (MED25) subunit of the eukaryotic Mediator complex [17,18,19,20]. This complex recruits to the MYCs-bound promoter all of the general transcription factors (GTFs) and RNA polymerase II machinery required to initiate JA transcriptional reprograming [19,20]. Repression of TFs by JAZ involves the competitive inhibition of MYC interaction with MED25 by the Jas domain of JAZs [17]. Repression also requires the recruitment of the general co-repressor TOPLESS (TPL) and TPL-related proteins through the NOVEL INTERACTOR OF JAZ (NINJA) adaptor, which are supposed to lock chromatin by histone deacetylation [21]. Under stress conditions, JA-Ile promotes the formation of JAZ-COI1 complexes, triggering JAZ ubiquitination and subsequent degradation via the 26S proteasome [9,10,11]. This leads to de-repression of the TFs that initiate the transcription of JA-dependent genes [22].
Pathogens have evolved capabilities to manipulate or subvert plant hormone signaling pathways to cause hormonal imbalances for their own benefit. Microbes enhance their virulence by producing plant hormones, phytohormone mimics, or injecting into eukaryotic plant cells an arsenal of virulence effector proteins that target hormonal components to evade or disrupt hormonal signaling pathways and/or crosstalk [23]. Here, we describe the molecular details of the best-characterized examples of specific JA hormonal manipulation by microbes, which exemplify the ingenious ways by which pathogens can take control over the plant’s hormone signaling network to suppress host immunity.
2. Production of JA Phytohormones and Mimics by Pathogens
Pathogens are capable of synthesizing phytohormones and phytohormone mimics to take control of the host immune system by inducing hormonal imbalances (Table 1). Probably, the best-studied example corresponds to coronatine (COR), a mimic of the bioactive JA-Ile hormone, produced by some strains of P. syringae [8,24,25]. COR, as JA-Ile, is perceived through the COI1/JAZ co-receptor complex, activating JA-dependent responses that, in turn, inhibit SA-dependent defenses required for P. syringae resistance (Figure 1) [5,26]. Remarkably, COR is more active than the JA-Ile plant hormone itself in triggering the COI1-JAZ complex formation and subsequent JAZ degradation, indicating that COR acts as a potent and highly specific mimic of JA-Ile perception in plants [8,27]. The functional similarity between COR and JA-Ile has been further demonstrated by the three-dimensional structural analysis of each molecule in association with a COI1 receptor complex [11], and by the rational design of COR-based antagonists of JA-Ile perception that block the COI1-JAZ co-receptor [28]. Among the twelve canonical JAZs, eight genes are induced in Arabidopsis during P. syringae infection in a COR-dependent manner [29]. Of note, only loss-of-function mutants of JAZ10 are more susceptible to P. syringae, indicating that JAZ10 is a negative regulator of both JA signaling and disease development [29]. In combination with jaz10, the jaz5 knockout (KO) enhances chlorotic symptoms and (moderately) bacterial growth, indicating that JAZ5 and JAZ10 act co-operatively to restrict P. syringae growth during bacterial pathogenesis and are targeted by COR [30].
In plants, COR acts as a multifunctional suppressor of defense responses and P. syringae mutants unable to produce COR show reduced virulence [24,31]. COR contributes to disease symptomatology by inducing chlorotic lesions [24,32,33], facilitates bacterial entry into the plant host by stimulating the re-opening of stomata after microbial-triggered stomatal closure [34,35,36], promotes bacterial growth by inhibiting SA-dependent defenses required for P. syringae resistance through the activation of its antagonistic JA pathway [37,38] and suppresses plant cell wall defense through perturbation of secondary metabolism [31]. Analyses of both plant and bacterial mutants indicate that COR elicits the JA pathway to repress SA-signaling, and that suppression of SA responses is a necessary component of the virulence action of COR [2,32,33,39,40,41]. A mechanism for suppression of SA-signaling by COR occurs though MYC2 TF that binds to the promoter and triggers the expression of the three NAC (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) ANAC19, ANAC55 and ANAC72 TFs [39]. These ANACs repress expression of genes involved in SA-biosynthesis, such as Isochorismate Synthase 2 (SID2), and induce BSMT1, a benzoic/salicylic acid carboxyl methyltransferase that reduces the biologically active pool of SA. Consequently, COR suppresses SA-mediated plant immunity against the bacteria, inducing stomatal reopening and bacterial propagation in both local and systemic tissues [39]. Remarkably, closely related NAC TFs, JA2 (Jasmonic Acid 2) and JA2L (JA2-like) differentially regulate stomatal closure and reopening during Pseudomonas attack in tomato [42], further indicating that the function of NAC TFs is conserved among different plant species.

Table 1.
Microorganisms able to synthetize jasmonates (JAs) and JA-mimics.
| Micro-Organism | Class | Molecules | Reference |
|---|---|---|---|
| Pseudomonas syringae | Pathogenic bacteria | coronatine | [25] |
| Lasiodiplodia theobromae | Pathogenic fungus | JA | [43] |
| Gibberella fujikuroi | Pathogenic fungus | JA-Ile and other jasmonates | [44] |
| Collybia confluens | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Collybia dryophila | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Coprinus alkalinus | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Coprinus cinereus | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Mycena tintinabulum | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Phellinus laevigatus | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Trametes versicolor | Saprophitic fungus | JA and 7-iso-JA | [45] |
| Pisolithus tinctorius | Ectomyccorhizal fungus | JA and 7-iso-JA | [46] |
| Fusarium oxysporum | Pathogenic fungus | JA, JA-Ile, 9,10-dihydro-JA and other jasmonates | [47,48,49] |
| Diplodia gossypina | Pathogenic fungus | JA and Me-JA | [50] |
| Strains SF2, SF3 and SF4 | Endophytic bacteria | OPDA and other jasmonates | [51] |
| Magnaporthe oryzae | Pathogenic fungus | JA, Me-JA and 12OH-JA | [52] |
A COI1-independent effect of COR has also been reported [31]. COR seems to suppress an SA-independent pathway contributing to callose deposition, a hallmark of plant resistance, by reducing accumulation of an indole glucosinolate via PEN2 (penetration 2)-dependent pathway [31]. However, this COI1-independent effect of COR awaits further confirmation due to the use of weak alleles of coi1. Thus, acquisition of COR by Pseudomonas pathogens has been of tremendous adaptive importance during host-pathogen evolution to manipulate the host hormonal network to promote susceptibility.
Several fungi such as Fusarium oxysporum (F. oxysporum) and Lasiodiplodia theobromae (L. theobromae) are also capable of producing bioactive JAs among others (Table 1, Figure 1) [23,43,44,45,46,47,50,51,53,54]. For example, the tomato infecting fungus F. oxysporum forma specialis (f. sp) lycopersici produces JAs through the specific lipoxygenase enzyme 13S-LOX, conserved with plant LOXs [48]. In addition, F. oxysporum f. sp. conglutinans and F. oxysporum f. sp. matthioli, which infect roots of Arabidopsis, produce JA-Ile and JA-Leu, respectively, in culture filtrates [49]. Consequently, JA-insensitivity suppresses infection by these F. oxysporum strains that produce detectable JA-Ile levels [49]. This indicates that several fungi have also evolved to produce JAs as a virulence strategy to promote infection [49].
On the other hand, some pathogens have evolved the competence to detoxify or inactivate compounds required for immunity including JAs [8,55]. For example, hydroxylation of JAs is a common catabolic mechanism contributing to switch-off JA signaling but producing derivatives that may have other functions [55]. The rice blast fungus Magnaporthe oryzae (M. oryzae) produces an antibiotic biosynthesis monooxygenase (Abm) that converts endogenous free JA into hydroxylated JA (12OH-JA) [52]. Both Abm and 12OH-JA are secreted during host penetration and help evade plant defenses [52]. Loss of Abm in M. oryzae leads to accumulation of plant-produced methyl (Me)JA blocking the invasive growth of the fungi, whereas exogenously added 12OH-JA markedly attenuates abmΔ-induced immunity in rice. Therefore, Abm might have a dual role, on one hand reducing the free JA, and therefore plant defenses, and, on the other hand, producing 12OH-JA, which facilitates M. oryzae infection [52].
3. Effector-Mediated Manipulation of JA Signaling Pathway by Bacterial and Oomycete Plant Pathogens
Bacterial pathogens such as P. syringae have evolved specific effectors to disrupt hormonal equilibrium by targeting JA signaling and/or JA-SA balance in the plant cell (Table 2, Figure 1) [6,56,57,58,59].
The Pseudomonas effectors HopX1 and HopZ1a activate the JA signaling pathway by targeting the JAZ repressors [56,57]. HopX1 from P. syringae pv. tabaci (Pta) 11528 encodes a cysteine protease that associates with JAZ proteins via its conserved ZIM domain and induces JAZ degradation in a proteasome- and COI1-independent manner, likely via its cysteine protease activity [56]. Ectopic expression of HopX1 in Arabidopsis induces the expression of JA-dependent genes and represses SA signaling. Delivery of HopX1 by the natural TTSS (Type Three Secretion System) of Pseudomonas partially re-opens stomata and promotes susceptibility during the infection process. Since Pta 11528 does not produce COR, HopX1 may have evolved as an alternative evolutionary strategy to manipulate core regulators of JA signaling.

Table 2.
Microbial effectors targeting JA-mediated defenses.
| Effector | Organism | Plant Target | Mode of Action | Reference |
|---|---|---|---|---|
| AvrB (MPK4) | P. syringae | RIN4/MPK4 | Activates JA defenses targeting | [58] |
| MPK4/RIN4/HSP90/RAR1 complex | ||||
| AvrB (AHA1) | P. syringae | RIN4/AHA1 | Activates JA defenses promoting AHA1-dependent | [59] |
| COI1/JAZ interaction and JAZ degradation | ||||
| HopZ1a | P. syringae | JAZs | Activates JA defenses promoting COI1-dependent | [57] |
| degradation of the JAZ repressors | ||||
| HopX1 | P. syringae | JAZs | Activates JA defenses promoting COI1-independent | [56] |
| degradation of the JAZ repressors | ||||
| RxL44 | H. arabidopsidis | MED19a | Induces MED19a degradation shifting the balance | [60] |
| from SA- to JA/ET-mediated defense | ||||
| MiSSP7 | L. bicolor | JAZs | Suppresses JA defenses stabilizing the JAZs | [61] |
| SSITL | S. sclerotiorum | unkown | Suppresses JA defenses | [62] |
| Exopolysaccharide | B. cinerea | unkown | Activates SA pathway to suppress JA defenses | [63] |
| Abm | M. oryzae | JA or unkown | Suppresses JA defenses by convertion of fungal JA | [52] |
| into 12OH-JA |
HopZ1a from P. syringae pv. syringae (Pss) strain A2 encodes a cysteine protease/acetyltransferase that acetylates several JAZ proteins interacting with their conserved Jas domain [57]. HopZ1a-mediated acetylation induces JAZ1 degradation through an undefined mechanism that is dependent on COI1, leading to the activation of JA-dependent gene expression, suppression of SA responses and plant susceptibility (Figure 1) [57]. Interestingly, this suggests that post-translational modifications of the Jas motif of JAZ1 may modulate the 26S proteasome degradation triggered by COI1 in the absence of the hormone. Indeed, post-translational modifications, including acetylation, have been shown to induce or repress proteasomal degradation [64]. Recently, the threonine residue T346 was identified as the main auto-acetylation site of HopZ1a by mass spectrometry, whereas two additional neighboring serine residues (S349 and S351) are required for the acetyltransferase activity of HopZ1a in vitro and indispensable for the virulence function of HopZ1a in planta [65,66]. These serine residues are important for the enzymatic activity of HopZ1a and are indispensable for inositol hexakisphosphate (IP6)-induced activation of the effector in eukaryotic hosts [65]. The absence of IP6 in bacteria suggests that the enzymatic activity of this effector is activated after it is delivered into the host. Interestingly, previous structural studies have revealed that interaction of the COI1/JAZ co-receptor with its JA-Ile ligand requires an inositol pentakisphosphate (IP5) cofactor [11]. Further investigations are needed to determine how HopZ1a-mediated acetylation of JAZs could facilitate their COI1-dependent degradation and whether this effector mimics a still unknown host post-translational mechanism controlling the activity of the JA receptor complex.
Besides the JA receptor, additional bacterial effectors affect plant hormonal equilibrium targeting other JA signaling components. The kinase-like AvrB effector was among the first effectors shown to promote JA signaling [58,67]. AvrB induces JA signaling by at least two different mechanisms, both dependent on the AvrB-interacting plasma membrane (PM)-associated protein RIN4 (RPM1-interacting) [68,69]. On one hand, AvrB interacts with and phosphorylates MPK4, a mitogen-activated protein kinase that acts as a positive regulator of JA signaling (Figure 1) [58]. AvrB is unable to elicit the expression of the plant defensin PDF1.2 in mpk4 mutant plants and when using an avrB mutant unable to phosphorylate MPK4, suggesting that phosphorylation of MPK4 is required for AvrB-mediated promotion of JA signaling [58]. Notably, overexpression of AvrB in the coi1 mutant background fails to enhance susceptibility to P. syringae [67], indicating that the AvrB-mediated defense suppression requires the JA receptor COI1 and canonical components of the JA signaling pathway.
On the other hand, AvrB also induces stomatal opening by positively regulating the PM H+-ATPase AHA1 to enhance bacterial penetration and virulence in a RIN4-dependent manner (Figure 1) [59,70,71]. RIN4 directly interacts with AHA1 and the closely related AHA2, acting as a regulator of the proton pump required for stomatal dynamics, likely to inhibit the entry of bacterial pathogens into the plant leaf during infection [71,72]. AvrB is able to interact with RIN4 and promote its phosphorylation [69,73,74], and RIN4 phosphorylation mimics show enhanced H+-ATPase activity and inhibited immune responses activated upon microbial perception in Arabidopsis [70]. Congruently, transgenic expression of AvrB enhances H+-ATPase activity of AHA1, and bacterially delivered AvrB induces stomatal opening and enhances infection in a RIN4-dependent manner, demonstrating that the RIN4-AHA1 pathway is exploited by AvrB for virulence [59]. Interestingly, AHA1 promotes the interaction between the JA receptor COI1 and JAZ proteins, activating JA signaling. Similarly, AvrB also induces the COI1-JAZ9 interaction and the degradation of multiple JAZ proteins [59]. Thus, AvrB-induced stomatal opening and virulence requires the canonical JA signaling pathway, which involves COI1, JAZs and also previously described ANAC19, ANAC55 and ANAC72 TFs [39,59]. The biochemical mechanism by which the PM AHA1 activity induces nuclear COI1-JAZ interaction remains unclear. One hypothesis speculates that activation of PM H+-ATPase could induce post-translational modification of JAZs leading to JAZ degradation as, for example, the acetyltransferase activity of the bacterial effector HopZ1a [57,59]. Alternatively, the activation of PM H+-ATPase could induce the generation of JAs or unknown signal molecules/cofactors such as inositol pentakisphosphate IP5 to potentiate the assembly of the COI1-JAZ complex [59]. In this context, a recent report suggests that inositol pyrophosphate IP8 may also be a critical cofactor in COI1-JAZ complex formation as Arabidopsis plants lacking VIH2, a diphosphoinositol pentakisphosphate kinase (PPIP5K) regulating the biosynthesis of IP8, are compromised in JA dependent responses [75]. In summary, these findings uncover a novel pathway exploited by AvrB that acts upstream of COI1 to regulate JA signaling and stomatal opening.
The nuclear effector RxL44 encoded by the oomycete downy mildew pathogen Hyaloperonospora arabidopsidis directly interacts with the mediator subunit MED19a (Mediator19a), resulting in the proteasome-mediated degradation of MED19a (Figure 1) [60]. The Mediator complex acts as a molecular bridge between transcriptional regulators and RNA polymerase II at the transcription start site, and has emerged as a key regulator of plant immunity [76]. MED25, another component of the Mediator complex, interacts with several TFs including MYC2 and MYC3, acting as an integrative hub for the regulation of JA-responsive gene expression [17,18]). RxL44-induced MED19a degradation shifts the balance of defense gene transcription from SA-responsive to JA/ET-mediated defense, enhancing susceptibility to biotrophs. In contrast with RxL44, plants overexpressing MED19a show significant higher levels of SA defenses [60]. Thus, RxL44 enhances susceptibility to biotrophs by attenuating SA-dependent gene expression.
Finally, recent data indicates that independently evolved virulence effectors converge onto hubs in a plant immune system network [77,78]. High throughput systematic protein-protein interaction approaches between proteins from the reference plant Arabidopsis thaliana and effector proteins from the bacteria P. syringae, the oomycete Hyaloperonospora arabidopsidis and the ascomycete Golovinomyces orontii pathogens, has allowed to develop a large-scale map of physical interactions [77,78]. These experiments yielded a map of more than 6000 interactions, resulting in a network that identifies host proteins onto which intraspecies and interspecies pathogen effectors converge [77,78]. Pathogens from different kingdoms deploy independently evolved virulence effectors that interact with a set of highly connected cellular hubs to facilitate their diverse life-cycle strategies, showing an interspecies effector convergence onto shared host proteins. Remarkably, some of the most significantly targeted plant hub proteins by effectors include immune- and hormone-related proteins such as JAZ3 and MED19a among others. Thus, it is tempting to speculate that the understanding of how effector molecules from multiple pathogens hijack hormonal pathways to promote disease is just beginning. Altogether, these examples indicate that the JA receptor complex stands as a major hub/target in the arms race between plants and pathogens.
4. Effector-Mediated Manipulation of JA Signaling Pathway by Necrotrophic Fungi
Compared to our knowledge on bacterial and oomycete effectors, the molecular mechanisms used by necrotrophic fungi to manipulate JA signaling are still very poorly understood. Infection by necrotrophs primarily relies on the production of nonspecific toxins and cell wall degrading enzymes. However, several examples suggest that this type of pathogens also manipulate hormonal pathways. The necrotrophic fungi B. cinerea secretes an exopolysaccharide (EPS) potentially acting as an elicitor of the SA pathway to suppress JA defenses in tomato plants (Table 2) [63]. In another recent example, the necrotrophic fungal pathogen Sclerotinia sclerotiorum secretes an effector-like protein called SSITL (sclerotinia sclerotiorum integrin-like) that suppresses JA-dependent defenses [62]. SSITL retains the conserved solvent-exposed Arg-Gly-Asp (RGD)-like motif of plant integrin-like proteins, such as NDR1 (nonrace-specific disease resistance1) involved in defense signaling; therefore suggesting that SSITL mimics the activity of this type of proteins [62]. Further studies are still needed to understand at the molecular level how these molecules act inside plant cells.
5. Effector-Mediated Modulation of JA Signaling by Beneficial Microbes
Beneficial microbes establish long-term relationships with their host plants to fulfill their life cycles [79]. Ectomycorrhizal fungi (EM), such as Laccaria bicolor, support host growth by providing growth-limiting nutrients to their plant host through a mutualistic symbiotic relationship within the plant roots [80]. In order to develop within the host and feed on living cells, they need to contend with the defense mechanisms of the plant, and fungal secreted effectors may facilitate this process. The mycorrhizal induced small secreted protein 7 (MiSSP7) is encoded by the most highly symbiosis-upregulated gene of the EM L. bicolor. MiSSP7 is an effector protein indispensable for the establishment of mutualism with their host plants [80]. Upon perception of diffusible signals from the Populus roots, the fungus secretes MiSSP7 that is imported into the plant cell via phosphatidylinositol-3-phosphate-mediated endocytosis, and targeted to the plant nucleus (Table 2, Figure 1) [61,80]. In the nucleus, MiSSP7 interacts with the JA-repressor PtJAZ6 and protects PtJAZ6 from JA-induced degradation. Consistently, the symbiotic activity of MiSSP7 can be replaced by either inhibiting JA signaling in planta or through transgenic overexpression of the PtJAZ6 gene [61]. This indicates that MiSSP7 is able to inhibit the JA-dependent host defenses stabilizing JAZ repressors to promote fungal colonization and mutualism. Thus, beneficial microbes also evolved effectors that are delivered inside the host plant cell to manipulate hormonal signaling, and thus can be considered symbiotic effectors.
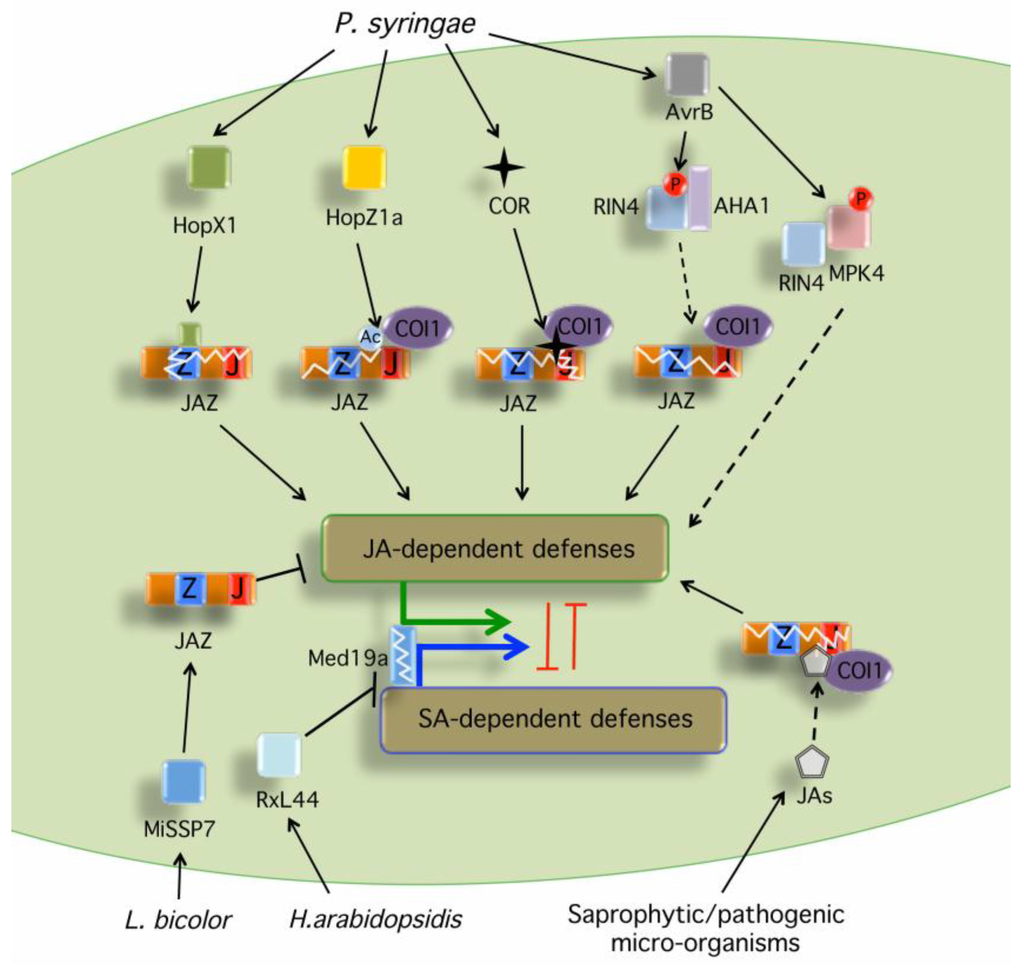
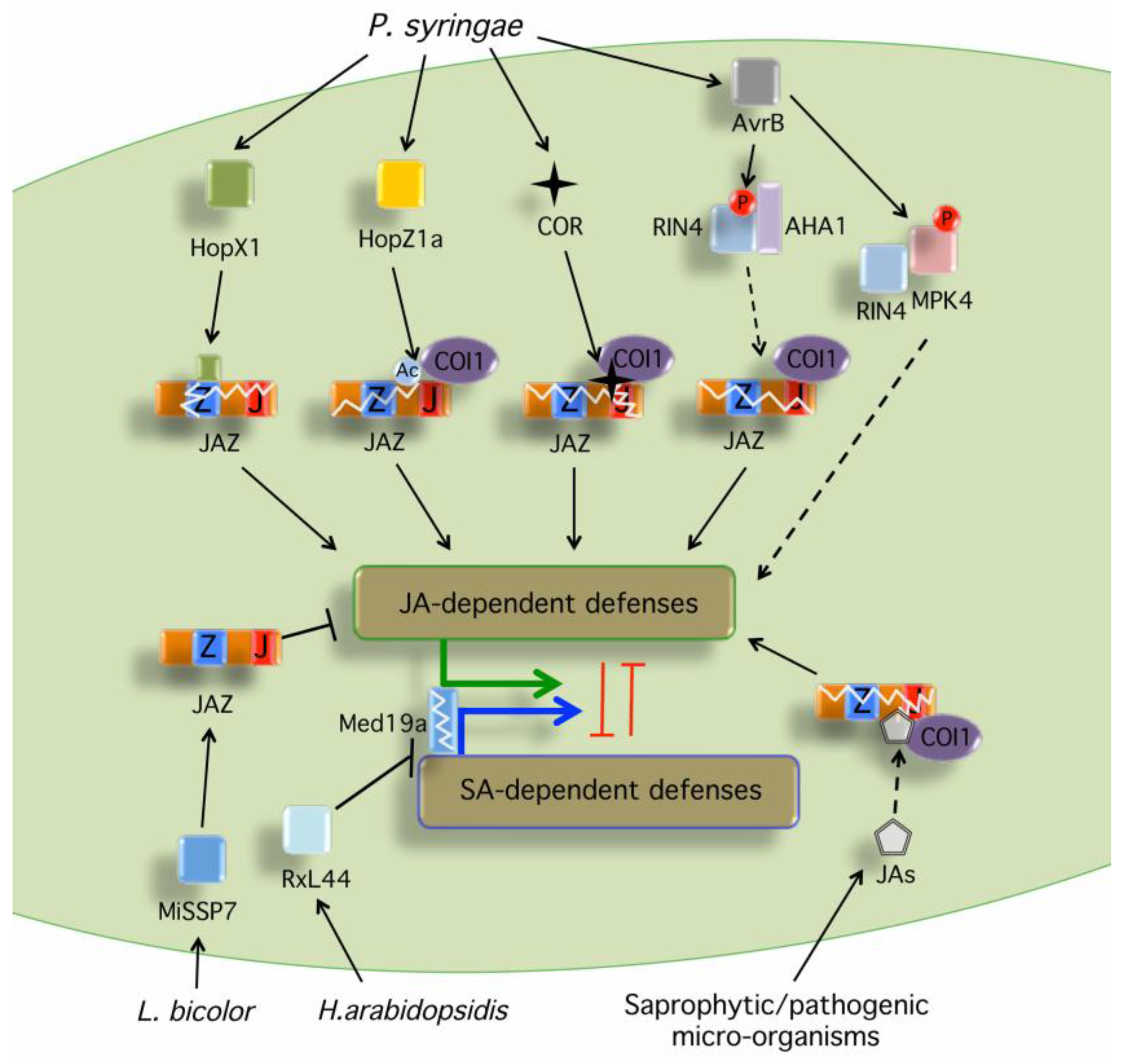
Figure 1.
Phytotoxins and microbial effectors targeting the JA signaling components. Several microbes produce JA-Ile precursors or the JA-Ile mimic coronatine that are perceived by the JA-Ile receptor complex and induce degradation of JAZ repressors (see Table 1 for specific examples). HopX1 from P. syringae pv. tabaci (Pta) 11528 encodes a cysteine protease that associates with JAZ proteins induces their degradation in a proteasome- and COI1-independent manner, likely via its cysteine protease activity [56]. HopZ1a from P. syringae pv. syringae (Pss) strain A2 encodes a cysteine protease/acetyltransferase that acetylates several JAZ proteins and induces their degradation through an undefined mechanism that is dependent on COI1 [57]. AvrB interacts with [71] and phosphorylates MPK4, which triggers activation of JA signaling [58]. AvrB also induces the degradation of multiple JAZ proteins by positively regulating the PM H+-ATPase AHA1 to enhance bacterial penetration and virulence through RIN4 [59]. The ectomycorrhizal MiSSP7 effector interacts with PtJAZ6 and protects PtJAZ6 from JA-induced degradation, attenuating JA-dependent host defenses to promote fungal colonization and mutualism [61]. The oomycete downy mildew effector RxL44 directly interacts with the mediator subunit MED19a (Mediator19a), resulting in the proteasome-mediated degradation of MED19a, which shifts the balance of defense gene transcription from SA-responsive to JA/ET-mediated defense, enhancing susceptibility to biotrophs [60]. In all cases, degradation of JAZs leads to de-repression of the TFs that initiate the activation of JA-dependent gene expression, suppression of SA responses and plant susceptibility. Black arrows indicate activation of the indicated hormonal pathway. Black bars indicate inhibition of the indicated hormonal pathway. Dashed arrows denote indirect or unclear mechanism leading to the activation of the indicated hormonal pathway. A circled Ac indicates acetylation. A circled P indicates phosphorylation.
Figure 1.
Phytotoxins and microbial effectors targeting the JA signaling components. Several microbes produce JA-Ile precursors or the JA-Ile mimic coronatine that are perceived by the JA-Ile receptor complex and induce degradation of JAZ repressors (see Table 1 for specific examples). HopX1 from P. syringae pv. tabaci (Pta) 11528 encodes a cysteine protease that associates with JAZ proteins induces their degradation in a proteasome- and COI1-independent manner, likely via its cysteine protease activity [56]. HopZ1a from P. syringae pv. syringae (Pss) strain A2 encodes a cysteine protease/acetyltransferase that acetylates several JAZ proteins and induces their degradation through an undefined mechanism that is dependent on COI1 [57]. AvrB interacts with [71] and phosphorylates MPK4, which triggers activation of JA signaling [58]. AvrB also induces the degradation of multiple JAZ proteins by positively regulating the PM H+-ATPase AHA1 to enhance bacterial penetration and virulence through RIN4 [59]. The ectomycorrhizal MiSSP7 effector interacts with PtJAZ6 and protects PtJAZ6 from JA-induced degradation, attenuating JA-dependent host defenses to promote fungal colonization and mutualism [61]. The oomycete downy mildew effector RxL44 directly interacts with the mediator subunit MED19a (Mediator19a), resulting in the proteasome-mediated degradation of MED19a, which shifts the balance of defense gene transcription from SA-responsive to JA/ET-mediated defense, enhancing susceptibility to biotrophs [60]. In all cases, degradation of JAZs leads to de-repression of the TFs that initiate the activation of JA-dependent gene expression, suppression of SA responses and plant susceptibility. Black arrows indicate activation of the indicated hormonal pathway. Black bars indicate inhibition of the indicated hormonal pathway. Dashed arrows denote indirect or unclear mechanism leading to the activation of the indicated hormonal pathway. A circled Ac indicates acetylation. A circled P indicates phosphorylation.
6. Conclusions
The importance of signaling molecules in plant immunity is a well-established notion in plant biology and the molecular mechanisms regulating the phytohormone-mediated defenses have been extensively studied in the last decades. In this context, the role of hormones in plant immunity is further highlighted by the increasing number of pathogens producing phytohormones or phytohormone mimics and by the identification of a plethora of microbial effectors targeting hormonal pathways to promote disease or establish beneficial interactions. Of note, virulence activities of effectors on specific hormonal components can lead to the identification of novel mechanisms of host hormonal regulation unknown to date. In addition, an improved mechanistic understanding of how pathogenic toxins and effectors manipulate host hormonal components will be instrumental in identifying new defensive hubs and to develop new plant varieties with improved resistance to pathogens.
Acknowledgments
We thank members of the Roberto Solano laboratory for critical reading of the manuscript. Selena Gimenez-Ibanez was supported by an International UNESCO-LÓREAL Fellowship for Women in Science. Andrea Chini was supported by a Ramon y Cajal fellowship from the Spanish Research Council. Work in Roberto Solano lab was funded by the Spanish Ministry for Science and Innovation grant BIO2013-44407-R.
Author Contributions
Selena Gimenez-Ibanez wrote the manuscript, which all authors read, edited and approved. Andrea Chini prepared the figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Solano, R. Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in annals of botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; van Wees, S.C. Hormonal modulation of plant immunity. Annu.Rev.Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Stolz, S.; Chetelat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St. Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-tify JAZ protein in arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Cevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. Mediator25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Kidd, B.N.; Edgar, C.I.; Kumar, K.K.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in arabidopsis. Plant Cell 2009, 21, 2237–2252. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Rowe, H.C.; Xiao, Y.; Chehab, E.W.; Kliebenstein, D.J.; Wagner, D.; Dehesh, K. The chromatin remodeler splayed regulates specific stress signaling pathways. PLoS Pathog. 2008, 4, e1000237. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. Ninja connects the co-repressor topless to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.M.; Hernandez-Guzman, G.; Kloek, A.P.; Alarcon-Chaidez, F.; Sreedharan, A.; Rangaswamy, V.; Penaloza-Vazquez, A.; Bender, C.L.; Kunkel, B.N. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of pseudomonas syringae pv. Tomato DC3000. Mol. Plant-Microbe Interact. 2004, 17, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Solano, R. Novel players fine-tune plant trade-offs. Essays Biochem. 2015, 58, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Monte, I.; Hamberg, M.; Chini, A.; Gimenez-Ibanez, S.; Garcia-Casado, G.; Porzel, A.; Pazos, F.; Boter, M.; Solano, R. Rational design of a ligand-based antagonist of jasmonate perception. Nat. Chem. Biol. 2014, 10, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of arabidopsis JAZ gene expression during pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2012, 13, 46–57. [Google Scholar] [CrossRef] [PubMed]
- De Torres Zabala, M.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin arabidopsis defence responses to pseudomonas syringae infection. New Phytol. 2015. [Google Scholar] [CrossRef]
- Geng, X.; Cheng, J.; Gangadharan, A.; Mackey, D. The coronatine toxin of pseudomonas syringae is a multifunctional suppressor of arabidopsis defense. Plant Cell 2012, 24, 4763–4774. [Google Scholar] [CrossRef] [PubMed]
- Uppalapati, S.R.; Ishiga, Y.; Wangdi, T.; Kunkel, B.N.; Anand, A.; Mysore, K.S.; Bender, C.L. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with pseudomonas syringae pv. Tomato DC3000. Mol. Plant Microbe Interact. 2007, 20, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Kloek, A.P.; Verbsky, M.L.; Sharma, S.B.; Schoelz, J.E.; Vogel, J.; Klessig, D.F.; Kunkel, B.N. Resistance to pseudomonas syringae conferred by an arabidopsis thaliana coronatine-insensitive (COI1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ishiga, Y.; Clermont, K.; Mysore, K.S. Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by nonhost bacterial pathogens. PeerJ 2013. [Google Scholar] [CrossRef] [PubMed]
- Laurie-Berry, N.; Joardar, V.; Street, I.H.; Kunkel, B.N. The arabidopsis thaliana jasmonate insensitive 1 gene is required for suppression of salicylic acid-dependent defenses during infection by pseudomonas syringae. Mol. Plant Microbe Interact. 2006, 19, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Bahrami, A.K.; Pringle, E.G.; Hernandez-Guzman, G.; Bender, C.L.; Pierce, N.E.; Ausubel, F.M. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 2005, 102, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of pseudomonas syringae. Planta 2014, 240, 1149–1165. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, D.C.; Galt, S.; Giles, D.; Turner, W.B. Metabolites of lasiodiplodia theobromae. J. Chem. Soc. C 1971, 5, 1623–1627. [Google Scholar] [CrossRef]
- Miersch, O.; Bruckner, B.; Schmidt, J.; Sembdner, G. Cyclopentane fatty acids from gibberella fujikuroi. Phytochemistry 1992, 31, 3835–3837. [Google Scholar] [CrossRef]
- Miersch, O.; Gunther, T.; Fritsche, W.; Sembdner, G. Jasmonates from different fungal species. Nat. Prod. Lett. 1993, 2, 293–299. [Google Scholar] [CrossRef]
- Miersch, O.; Regvar, M.; Wasternack, C. Metabolism of jasmonic acid in pisolithus tinctorius cultures. Phyton 1999, 39, 243–247. [Google Scholar]
- Miersch, O.; Bohlmann, H.; Wasternack, C. Jasmonates and related compounds from fusarium oxysporum. Phytochemistry 1999, 50, 517–523. [Google Scholar] [CrossRef]
- Brodhun, F.; Cristobal-Sarramian, A.; Zabel, S.; Newie, J.; Hamberg, M.; Feussner, I. An iron 13S-lipoxygenase with an alpha-linolenic acid specific hydroperoxidase activity from fusarium oxysporum. PLoS ONE 2013, 8, e64919. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.J.; Yoon, A.J.; Faull, K.F.; Diener, A.C. Host perception of jasmonates promotes infection by fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 2014, 15, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Go, I.H.; Kim, K.J.; Kim, Y.H. Optimal conditions for the production of (+)-Jasmonic acid by Diplodia gossypina ATCC10936. Korean J. Microbiol. 2006, 42, 210–215. [Google Scholar]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Patkar, R.N.; Benke, P.I.; Qu, Z.; Constance Chen, Y.Y.; Yang, F.; Swarup, S.; Naqvi, N.I. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat. Chem. Biol. 2015, 11, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Miersch, O.; Porzel, A.; Wasternack, C. Microbial conversion of jasmonates-hydroxylations by aspergillus niger. Phytochemistry 1999, 50, 1147–1152. [Google Scholar] [CrossRef]
- Tsukada, K.; Takahashi, K.; Nabeta, K. Biosynthesis of jasmonic acid in a plant pathogenic fungus, lasiodiplodia theobromae. Phytochemistry 2010, 71, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Miersch, O.; Neumerkel, J.; Dippe, M.; Stenzel, I.; Wasternack, C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2008, 177, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Fernandez-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yao, J.; Ma, K.W.; Zhou, H.; Song, J.; He, S.Y.; Ma, W. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Xue, L.; Chu, J.; Yan, C.; Fu, J.; Chen, M.; Innes, R.W.; Zhou, J.M. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP KINASE 4. Cell Host Microbe 2010, 7, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.M. An arabidopsis plasma membrane proton atpase modulates JA signaling and is exploited by the pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C.; Asai, S.; Rallapalli, G.; Piquerez, S.; Fabro, G.; Jones, J.D. A downy mildew effector attenuates salicylic acid-triggered immunity in arabidopsis by interacting with the host mediator complex. PLoS Biol. 2013, 11, e1001732. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssieres, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MISSP7 of the mutualistic fungus laccaria bicolor stabilizes the populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wei, W.; Fu, Y.; Cheng, J.; Xie, J.; Li, G.; Yi, X.; Kang, Z.; Dickman, M.B.; Jiang, D. A secretory protein of necrotrophic fungus sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 2013, 8, e53901. [Google Scholar] [CrossRef] [PubMed]
- El Oirdi, M.; el Rahman, T.A.; Rigano, L.; el Hadrami, A.; Rodriguez, M.C.; Daayf, F.; Vojnov, A.; Bouarab, K. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 2011, 23, 2405–2421. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Bae, M.K.; Ahn, M.Y.; Kim, S.H.; Sohn, T.K.; Bae, M.H.; Yoo, M.A.; Song, E.J.; Lee, K.J.; Kim, K.W. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef]
- Ma, K.W.; Jiang, S.; Hawara, E.; Lee, D.; Pan, S.; Coaker, G.; Song, J.; Ma, W. Two serine residues in pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 2015, 208, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Rufian, J.S.; Lucia, A.; Macho, A.P.; Orozco-Navarrete, B.; Arroyo-Mateos, M.; Bejarano, E.R.; Beuzon, C.R.; Ruiz-Albert, J. Auto-acetylation on K289 is not essential for HopZ1a-mediated plant defense suppression. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, X.; Cui, H.; He, P.; Thilmony, R.; Chintamanani, S.; Zwiesler-Vollick, J.; Gopalan, S.; Tang, X.; Zhou, J.M. RAR1, a central player in plant immunity, is targeted by pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 2006, 103, 19200–19205. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Godiard, L.; Straube, E.; Ashfield, T.; Lewald, J.; Sattler, A.; Innes, R.W.; Dangl, J.L. Structure of the arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 1995, 269, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Mackey, D.; Holt, B.F., 3rd; Wiig, A.; Dangl, J.L. RIN4 interacts with pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef]
- Lee, D.; Bourdais, G.; Yu, G.; Robatzek, S.; Coaker, G. Phosphorylation of the plant immune regulator rpm1-interacting protein4 enhances plant plasma membrane h+-atpase activity and inhibits flagellin-triggered immune responses in arabidopsis. Plant Cell 2015, 27, 2042–2056. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Elmore, J.M.; Fuglsang, A.T.; Palmgren, M.G.; Staskawicz, B.J.; Coaker, G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009, 7, e1000139. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.M.; Coaker, G. The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol. Plant 2011, 4, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.H.; da Cunha, L.; Wu, A.J.; Gao, Z.; Cherkis, K.; Afzal, A.J.; Mackey, D.; Dangl, J.L. Specific threonine phosphorylation of a host target by two unrelated type iii effectors activates a host innate immune receptor in plants. Cell Host Microbe 2011, 9, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Elmore, J.M.; Lin, Z.J.; Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target rin4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011, 9, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Johnen, P.; Azevedo, C.; Dynowski, M.; Weiss, M.; Capolicchio, S.; Mao, H.; Iven, T.; Steenbergen, M.; Freyer, M.; et al. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in arabidopsis. Plant Cell 2015, 27, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Conaway, R.C.; Conaway, J.W. Function and regulation of the mediator complex. Curr. Opin. Genet. Dev. 2011, 21, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Wessling, R.; Epple, P.; Altmann, S.; He, Y.; Yang, L.; Henz, S.R.; McDonald, N.; Wiley, K.; Bader, K.C.; Glasser, C.; et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 2014, 16, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legue, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
