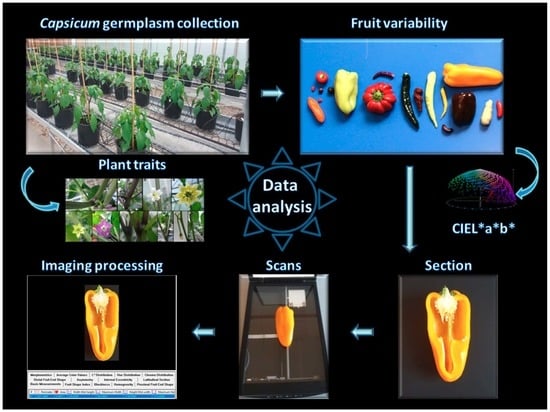
Large Scale Phenotyping Provides Insight into the Diversity of Vegetative and Reproductive Organs in a Wide Collection of Wild and Domesticated Peppers (Capsicum spp.)
Abstract
1. Introduction
2. Results
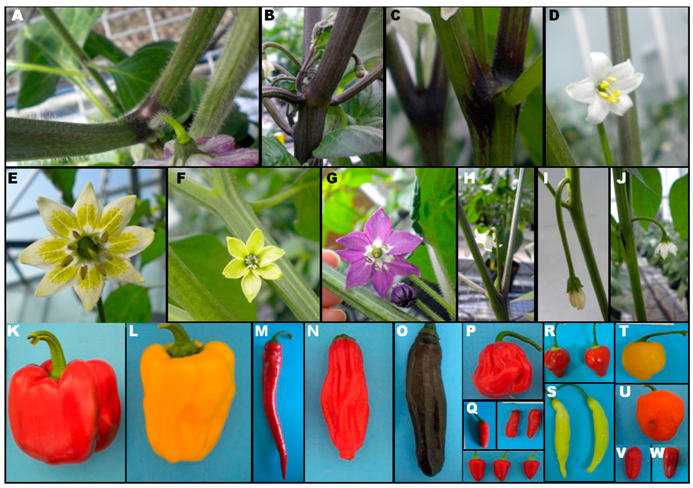
2.1. Diversity among Accessions
2.2. Diversity between Capsicum Species
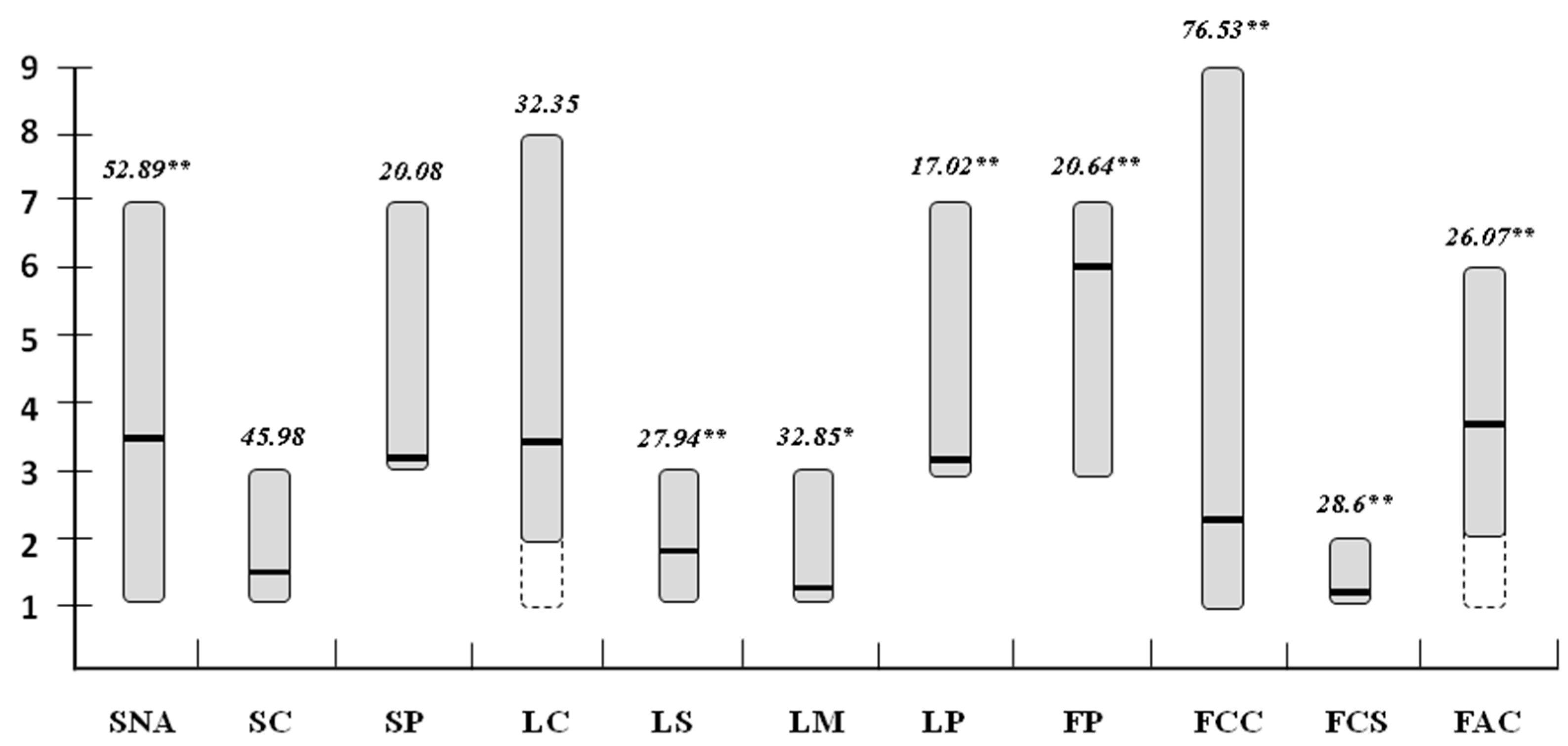
2.3. Diversity between Sweet and Hot Cultivated Pepper Types
2.4. Multivariate Analyses
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Phenotypic Characterization
4.3. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Carrizo García, C.; Barfuss, M.H.J.; Sehr, E.M.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 2016, 118, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, B. Genetic resources and breeding of Capsicum spp. Euphytica 1997, 96, 129–133. [Google Scholar] [CrossRef]
- Bosland, P.W. Capsicum Pepper: Varieties and Classification. Available online: http://contentdm.nmsu.edu/cdm/ref/collection/AgCircs/id/12518 (accessed on 18 November 2018).
- Paran, I.; Fallik, E. Breeding for fruit quality in pepper (Capsicum spp). In Breeding for Fruit Quality; Jenks, M.A., Bebeli, P.J., Eds.; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Albrecht, E.; Zhang, D.; Saftner, R.A.; Stommel, J.R. Genetic diversity and population structure of Capsicum baccatum genetic resources. Genet. Resour. Crop Evol. 2012, 59, 517–538. [Google Scholar] [CrossRef]
- Nicolaï, M.; Cantet, M.; Lefebvre, V.; Sage-Palloix, A.M.; Palloix, A. Genotyping a large collection of pepper (Capsicum spp) with SSR loci brings new evidence for the wild origin of cultivated C annuum and the structuring of genetic diversity by human selection of cultivar types. Genet. Resour. Crop. Evol. 2013, 60, 2375–2390. [Google Scholar] [CrossRef]
- Gonzalez-Perez, S.; Garces-Claver, A.; Mallor, C.; Saenz de Miera, L.E.; Fayos, O.; Pomar, F.; Merino, F.; Silvar, C. New insights into Capsicum spp relatedness and the diversification process of Capsicum annuum in Spain. PLoS ONE 2014, 9, e116276. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.; Umaharan, P.; Dayanandan, S. Microsatellite based analysis of the genetic structure and diversity of Capsicum chinense in the Neotropics. Genet. Resour. Crop Evol. 2014, 61, 741–755. [Google Scholar] [CrossRef]
- Lee, H.Y.; Ro, N.Y.; Jeong, H.J.; Kwon, J.K.; Jo, J.; Ha, Y.; Jung, A.; Han, J.W.; Venkatesh, J.; Kang, B.C. Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet. 2016, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Taranto, F.; D’Agostino, N.; Greco, B.; Cardi, T.; Tripodi, P. Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotype-by-sequencing. BMC Genom. 2016, 17, 943. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, P.; Massa, D.; Venezia, A.; Cardi, T. Sensing Technologies for Precision Phenotyping in Vegetable Crops: Current Status and Future Challenges. Agronomy 2018, 8, 54. [Google Scholar] [CrossRef]
- Bioversity International. Available online: https://www.bioversityinternational.org/e-library/publications/descriptors/ (accessed on 18 October 2018).
- Brewer, M.T.; Moyseenko, J.B.; Monforte, A.J.; van der Knaap, E. Morphological variation in tomato: A comprehensive study of quantitative trait loci controlling fruit shape and development. J. Exp. Bot. 2007, 58, 1339–1349. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, M.J.; van der Knaap, E. A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape. Theor. Appl. Genet. 2008, 116, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, M.J.; Brewer, M.T.; Anderson, C.; Sullivan, D.; Gray, S.; van der Knaap, E. Tomato fruit shape analysis using morphometric and morphology attributes implemented in Tomato Analyzer software program. J. Am Soc. Hortic. Sci. 2009, 134, 77–87. [Google Scholar]
- Mazzucato, A.; Ficcadenti, N.; Caioni, M.; Mosconi, P.; Piccinini, P.; Sanampudi, V.R.R.; Sestili, S.; Ferrari, V. Genetic diversity and distinctiveness in tomato (Solanum lycopersicum L.) landraces: The Italian case study of ‘A pera Abruzzese’. Sci. Hort. 2010, 125, 55–62. [Google Scholar] [CrossRef]
- Rodriguez, G.R.; Kim, H.J.; van der Knaap, E. (2013) Mapping of two suppressors of OVATE (sov) loci in tomato. Heredity 2013, 111, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.R.; Munoz, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Mc Spadden Gardener, B.B.; Francis, D.; van der Knaap, E. Distribution of SUN OVATE LC and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Figàs, M.R.; Prohens, J.; Raigón, M.D.; Fernández-de-Córdova, P.; Fita, A.; Soler, S. Characterization of a collection of local varieties of tomato (Solanum lycopersicum L) using conventional descriptors and the high-throughput phenomics tool Tomato Analyzer. Genet. Resour. Crop Evol. 2015, 62, 189–204. [Google Scholar] [CrossRef]
- Plazas, M.; Andújar, I.; Vilanova, S.; Gramazio, P.; Herraiz, F.J.; Prohens, J. Conventional and phenomics characterization provides insight into the diversity and relationships of hypervariable scarlet (Solanum aethiopicum L) and gboma (S macrocarpon L) eggplant complexes. Front. Plant Sci. 2014, 5, 318. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, P.; Prohens, J.; Vilanova, S.; Gramazio, P.; Plazas, M. Phenotyping of Eggplant Wild Relatives and Interspecific Hybrids with Conventional and Phenomics Descriptors Provides Insight for Their Potential Utilization in Breeding. Front. Plant Sci. 2016, 7, 677. [Google Scholar] [CrossRef] [PubMed]
- Naegele, R.P.; Mitchell, J.; Hausbeck, M.K. Genetic Diversity Population Structure and Heritability of Fruit Traits in Capsicum annuum. PLoS ONE 2016, 11, e0156969. [Google Scholar] [CrossRef] [PubMed]
- Yarnes, S.C.; Ashrafi, H.; Reyes-Chin-Wo, S.; Hill, T.A.; Stoffel, K.M.; Van Deynze, A. Identification of QTLs for capsaicinoids fruit quality and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 2013, 56, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.A.; Chunthawodtiporn, J.; Afshrafi, H.; Stoffel, K.; Weir, A.; Van Deynze, A. Region underlying population structure and the genomics of organ size determination in Capsicum annuum. Plant Genome 2017, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Gould, K.S. Nature’s Swiss Army Knife, The Diverse Protective Roles of Anthocyanins in Leaves. J. Biomed. Biotechnol. 2005, 5, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Gould, K.S.; Dudle, D.A.; Neufeld, H.S. Why some stems are red, cauline anthocyanins shield photosystem II against high light stress. J. Exp. Bot. 2010, 61, 2707–2717. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.T.; Gurr, G.M. Trichomes of Lycopersicon species and their hybrids, Effects on pests and natural enemies. Agr. Forest Entomol. 2005, 7, 265–276. [Google Scholar] [CrossRef]
- Di Dato, F.; Parisi, M.; Cardi, T.; Tripodi, P. Genetic diversity and assessment of markers linked to resistance and pungency genes in Capsicum germplasm. Euphytica 2015, 1, 103–119. [Google Scholar] [CrossRef]
- Cremona, S.; Iovene, M.; Festa, G.; Conicella, C.; Parisi, M. Production of embryo rescued hybrids between the landrace ‘‘Friariello’’ (Capsicum annuum var annuum) and C baccatum var pendulum, phenotypic and cytological characterization. Euphytica 2018, 214, 129. [Google Scholar] [CrossRef]
- Paran, I.; der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 2007, 58, 3841–3852. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, M.; Vilanova, S.; Plazas, M.; Gramazio, P.; Herraiz, F.J.; Andújar, I.; Prohens, J. Phenomics of fruit shape in eggplant (Solanum melongena L) using tomato Analyzer software. Sci. Hort. 2013, 164, 625–632. [Google Scholar] [CrossRef]
- Thorup, T.A.; Tanyolac, B.; Livingstone, K.D.; Popovsky, S.; Paran, I.; Jahn, M. Candidate gene analysis of organ pigentation loci in the Solanaceae. Proc. Natl. Acad. Sci. USA 2000, 97, 11192–11197. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; Kuntz, M.; Camara, B.; Palloix, A. The capsanthin-capsorubin synthase gene, a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol. Biol. 1998, 36, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Ben Chaim, A.; Borovsky, E.; Rao, G.U.; Tanyolac, B.; Paran, I. fs31, a major fruit shape QTL conserved in Capsicum. Genome 2003, 46, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zygier, S.; Ben Chaimm, A.; Efrati, A.; Kaluzky, G.; Borovsky, Y.; Paran, I. QTLs mapping for fruit size and shape in chromosomes 2 and 4 in pepper and a comparison of the pepper QTL map with that of tomato. Theor. Appl. Genet. 2005, 111, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Barchi, L.; Lefebvre, V.; Sage-Palloix, A.M.; Lanteri, S.; Palloix, A. QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 2009, 118, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, E.; Paran, I. Characterization of fs101 a major QTL controlling fruit elongation in Capsicum. Theor. Appl. Genet. 2011, 123, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jeong, H.J.; Yang, H.B.; Kang, S.M.; Kwon, J.K.; Kim, S.; Choi, D.; Kang, B.C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Nair, N.; Sreenivasan, T. A method for establishing a core collection of Saccharum officinarum L germplasm based on quantitative-morphological data. Genet. Resour. Crop Evol. 2000, 47, 1–9. [Google Scholar] [CrossRef]
- Tomato Analyzer manual. Available online: https://vanderknaaplab.uga.edu/files/Tomato_Analyzer_4_Manual.pdf (accessed on 2 November 2018).
- Curtin, F.; Schulz, P. Multiple correlations and Bonferroni’s correction. Biol. Psychiatry 1998, 44, 775–777. [Google Scholar]
- Basu, S.; Duren, W.; Evans, C.R.; Burant, C.; Michailidis, G.; Karnovsky, A. Sparse network modeling and Metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 2017, 33, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Tarcea, V.G.; Karnovsky, A.; Mire, L.B.R.; Weymouth, T.E.; Beecher, C.W.; Cavalcoli, J.D.; Athey, B.D.; Omenn, G.S.; Burant, C.F.; et al. Metscape, a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 2010, 26, 971–973. [Google Scholar] [CrossRef] [PubMed]





| Traits | Acronim | MS a | TSS% | R | F Value b | Mean | Range | CV |
|---|---|---|---|---|---|---|---|---|
| Fruit descriptors | ||||||||
| Perimeter | [P] | 3880.00 | 3.44 | 0.94 | 467.85 * | 18.99 | 67.45–2.06 | 60.53 |
| Area | [A] | 6222.19 | 5.52 | 0.89 | 248.87 * | 15.10 | 133.77–0.26 | 98.98 |
| Width Mid-height | [WMH] | 88.61 | 0.08 | 0.94 | 462.31 * | 2.51 | 15.91–0.41 | 69.24 |
| Maximum Width | [MW] | 102.74 | 0.09 | 0.89 | 236.49 * | 3.15 | 18.33–0.49 | 61.13 |
| Height Mid-width | [HMW] | 373.94 | 0.33 | 0.85 | 169.69 * | 5.92 | 22.9–0.42 | 63.48 |
| Maximum Height | [MH] | 532.37 | 0.47 | 0.93 | 417.51 * | 6.72 | 26.34–0.68 | 63.58 |
| Curved Height | [CH] | 566.22 | 0.50 | 0.94 | 457.78 * | 7.11 | 26.88–0.8 | 61.82 |
| Fruit Shape Index External I | [FSEI] | 37.01 | 0.03 | 0.76 | 95.73 * | 2.34 | 12.52–0.37 | 53.31 |
| Fruit Shape Index External II | [FSEII] | 104.25 | 0.09 | 0.74 | 87.64 * | 2.91 | 25.76–0.2 | 72.8 |
| Curved Fruit Shape Index | [FSC] | 194.28 | 0.17 | 0.87 | 210.26 * | 3.53 | 25.07–0.42 | 75.49 |
| Proximal Fruit Blockiness | [PFB] | 1.17 | 0.00 | 0.55 | 36.78 * | 0.90 | 3.23–0.03 | 28.92 |
| Distal Fruit Blockiness | [DFB] | 0.42 | 0.00 | 0.27 | 11.16 * | 0.58 | 5.04–0.02 | 38.77 |
| Fruit Shape Triangle | [FST] | 7.54 | 0.01 | 0.17 | 6.39 * | 1.77 | 56.5–0.04 | 66.58 |
| Ellipsoid | [E] | 0.04 | 0.00 | 0.64 | 54.85 * | 0.09 | 0.27–0.01 | 47.2 |
| Circular | [C] | 0.47 | 0.00 | 0.92 | 345.14 * | 0.26 | 0.5–0.02 | 48.15 |
| Rectangular | [R] | 0.19 | 0.00 | 0.51 | 31.83 * | 0.41 | 0.72–0.02 | 27.00 |
| Shoulder Height | [SH] | 0.01 | 0.00 | 0.16 | 5.74 * | 0.02 | 0.24–0 | 136.35 |
| Proximal Angle Micro | [PMI] | 16,146.46 | 14.33 | 0.07 | 2.15 * | 127.01 | 359.2–0 | 69.42 |
| Proximal Angle Macro | [PMA] | 25,968.55 | 23.05 | 0.18 | 6.45 * | 102.64 | 353.8–0 | 67.01 |
| Proximal Indentation Area | [PIA] | 0.01 | 0.00 | 0.15 | 5.29 * | 0.02 | 0.67–0 | 201.39 |
| Distal Angle Micro | [DMI] | 18,696.80 | 16.60 | 0.11 | 3.72 * | 107.43 | 359.7–0 | 68.78 |
| Distal Angle Macro | [DMA] | 28,587.58 | 25.38 | 0.23 | 9.05 * | 83.39 | 357.2–0 | 75.58 |
| Distal Indentation Area | [DIA] | 0.01 | 0.00 | 0.19 | 7.30 * | 0.01 | 1.51–0 | 321.27 |
| Distal End Protrusion | [DEA] | 0.32 | 0.00 | 0.15 | 5.47 * | 0.13 | 9.62–0 | 193.1 |
| Obovoid | [OB] | 0.05 | 0.00 | 0.21 | 8.02 * | 0.02 | 1.01–0 | 360.61 |
| Ovoid | [OV] | 0.51 | 0.00 | 0.53 | 34.21 * | 0.30 | 0.89–0 | 58.86 |
| V. Asymmetry | [ASv] | 2.24 | 0.00 | 0.58 | 41.23 * | 0.29 | 6.41–0 | 123.48 |
| H. Asymmetry.ob | [ASob] | 0.55 | 0.00 | 0.17 | 6.04 * | 0.06 | 6.15–0 | 556.95 |
| H. Asymmetry.ov | [Asov] | 17.47 | 0.02 | 0.66 | 58.39 * | 0.77 | 7.83–0 | 119.57 |
| Width Widest Pos | [WWP] | 0.30 | 0.00 | 0.35 | 16.31 * | 0.35 | 0.99–0.02 | 47.22 |
| Eccentricity | [EC] | 0.11 | 0.00 | 0.32 | 14.39 * | 0.72 | 0.8–0.02 | 14.25 |
| Proximal Eccentricity | [PEC] | 0.18 | 0.00 | 0.05 | 1.75 * | 0.90 | 24.67–0.02 | 36.27 |
| Distal Eccentricity | [DEC] | 5.95 | 0.01 | 0.07 | 2.09 * | 0.93 | 166–0.14 | 184.06 |
| Fruit Shape Index Internal | [FSI] | 106.52 | 0.09 | 0.73 | 80.38 * | 2.94 | 26.35–0.2 | 73.65 |
| Eccentricity Area Index | [EA] | 0.15 | 0.00 | 0.46 | 25.33 * | 0.45 | 0.99–0 | 22.47 |
| Lobedness Degree | [LD] | 11,008.87 | 9.77 | 0.81 | 129.89 * | 27.12 | 189.84–1.07 | 76.81 |
| Pericarp Area | [PA] | 0.03 | 0.00 | 0.42 | 21.86 * | 0.57 | 1.22–0.54 | 8.06 |
| Pericarp Thickness | [PT] | 0.01 | 0.00 | 0.51 | 31.22 * | 0.22 | 0.26–0.05 | 11.51 |
| Color descriptors | ||||||||
| L * | [L] | 1673.04 | 9.47 | 0.87 | 100.39 * | 42.50 | 97.65–13.31 | 26.18 |
| a * | [a] | 1733.39 | 9.81 | 0.85 | 82.84 * | 27.65 | 58.33–(−19.61) | 41.50 |
| b * | [b] | 3851.92 | 21.80 | 0.93 | 185.38 * | 27.15 | 95.72–(−9.48) | 60.36 |
| Chroma | [CHR] | 2746.17 | 15.54 | 0.88 | 102.13 * | 41.22 | 96.88–0.62 | 34.55 |
| Hue angle | [HA] | 7666.55 | 43.38 | 0.75 | 42.55 * | 35.89 | 89.90–(−89.98) | 71.80 |
| Plant Descriptors | C. Annuum Sweet (n = 45) | C. Annuum Hot (n = 135) | Prob > F | ||||
|---|---|---|---|---|---|---|---|
| Mean | Range | CV | Mean | Range | CV | ||
| [SNA] | 3.86 | 7–1 | 42.25 | 4.17 | 7–1 | 37.40 | 0.336 |
| [SC] | 1.42 | 3–1 | 40.60 | 1.49 | 3–1 | 43.93 | 0.635 |
| [SP] | 3.00 | 3–3 | 0.00 | 3.21 | 7–3 | 29.90 | 0.113 |
| [LC] | 3.00 | 3–3 | 0.00 | 3.50 | 8–2 | 37.73 | 0.042 |
| [LS] | 1.90 | 3–1 | 17.40 | 2.00 | 3–1 | 24.60 | 0.732 |
| [LM] | 1.10 | 2–1 | 29.00 | 1.10 | 2–1 | 29.00 | 0.969 |
| [LP] | 3.00 | 3–3 | 0.00 | 3.10 | 7–3 | 21.60 | 0.259 |
| [FP] | 6.00 | 7–3 | 20.30 | 5.80 | 7–3 | 24.60 | 0.323 |
| [FCC] | 1.30 | 2–1 | 35.60 | 2.10 | 9–1 | 99.00 | 0.022 |
| [FCS] | 1.01 | 2–1 | 16.70 | 1.02 | 2–1 | 11.20 | 0.553 |
| [FAC] | 3.91 | 5–2 | 14.36 | 4.01 | 5–2 | 13.70 | 0.384 |
| Fruit descriptors | |||||||
| [P] | 33.58 | 66.58–6.39 | 26.72 | 20.24 | 67.45–3.57 | 58.89 | <0.001 |
| [A] | 42.27 | 133.77–0.36 | 41.36 | 14.27 | 72.54–0.36 | 86.37 | <0.001 |
| [WMH] | 6.06 | 15.91–0.62 | 36.23 | 2.06 | 7.7–0.46 | 49.23 | <0.001 |
| [MW] | 6.85 | 18.33–0.71 | 29.95 | 2.91 | 13.17–0.71 | 49.34 | <0.001 |
| [HMW] | 8.79 | 21.32–0.67 | 39.83 | 6.55 | 22.9–0.42 | 64.41 | <0.001 |
| [MH] | 9.90 | 22.19–1.02 | 36.72 | 7.63 | 26.34–0.94 | 63.11 | <0.001 |
| [CH] | 10.89 | 22.59–2.09 | 33.56 | 7.96 | 26.88–1.2 | 61.47 | <0.001 |
| [FSEI] | 1.60 | 5.05–0.37 | 50.43 | 2.73 | 12.52–0.37 | 51.32 | <0.001 |
| [FSEII] | 1.80 | 12.99–0.29 | 74.17 | 3.65 | 25.76–0.2 | 70.58 | <0.001 |
| [FSC] | 2.22 | 15.97–0.42 | 74.45 | 4.54 | 25.07–0.42 | 71.79 | <0.001 |
| [PFB] | 0.88 | 2.3–0.08 | 30.83 | 0.99 | 2.76–0.03 | 27.48 | <0.001 |
| [DFB] | 0.61 | 3.21–0.08 | 37.43 | 0.60 | 5.04–0.04 | 41.91 | 0.018 |
| [FST] | 1.58 | 11.79–0.05 | 48.76 | 1.85 | 20.89–0.05 | 56.57 | <0.001 |
| [E] | 0.09 | 0.23–0.01 | 40.85 | 0.10 | 0.27–0.01 | 47.91 | <0.001 |
| [C] | 0.20 | 0.48–0.02 | 52.01 | 0.30 | 0.5–0.02 | 42.58 | <0.001 |
| [R] | 0.48 | 0.72–0.05 | 20.05 | 0.39 | 0.7–0.02 | 30.70 | <0.001 |
| [SH] | 0.03 | 0.24–0 | 109.88 | 0.03 | 0.19–0 | 132.82 | <0.001 |
| [PMI] | 145.56 | 357.8–0 | 69.86 | 122.20 | 359.2–0 | 72.94 | <0.001 |
| [PMA] | 133.53 | 352.1–0 | 67.57 | 96.83 | 353.8–0 | 71.95 | <0.001 |
| [PIA] | 0.04 | 0.67–0 | 190.88 | 0.02 | 0.35–0 | 160.20 | <0.001 |
| [DMI] | 127.60 | 356.7–0.2 | 64.50 | 104.79 | 359.7–0 | 68.31 | <0.001 |
| [DMA] | 99.24 | 351.7–0 | 67.05 | 79.03 | 357.2–0 | 84.98 | <0.001 |
| [DIA] | 0.02 | 0.66–0 | 264.11 | 0.01 | 1.51–0 | 401.60 | <0.001 |
| [DEA] | 0.09 | 1.15–0 | 216.62 | 0.14 | 9.62–0 | 218.36 | <0.001 |
| [OB] | 0.03 | 0.71–0 | 306.91 | 0.02 | 1.01–0 | 484.94 | 0.015 |
| [OV] | 0.27 | 0.82–0 | 64.86 | 0.36 | 0.89–0 | 48.62 | <0.001 |
| [ASv] | 0.42 | 6.41–0.02 | 118.12 | 0.34 | 4.53–0.01 | 119.48 | <0.001 |
| [ASob] | 0.07 | 5.38–0 | 509.16 | 0.05 | 6.15–0 | 716.74 | 0.15 |
| [Asov] | 0.81 | 6.91–0 | 101.32 | 1.05 | 7.83–0 | 108.57 | <0.001 |
| [WWP] | 0.35 | 0.9–0.04 | 44.15 | 0.30 | 0.99–0.02 | 55.78 | <0.001 |
| [EC] | 0.71 | 0.8–0.12 | 12.16 | 0.70 | 0.8–0.02 | 18.51 | 0.29 |
| [PEC] | 0.89 | 3.61–0.14 | 12.37 | 0.90 | 12.31–0.02 | 35.58 | 0.28 |
| [DEC] | 0.90 | 5.51–0.5 | 17.68 | 0.98 | 166–0.18 | 263.60 | 0.33 |
| [FSI] | 1.81 | 13.15–0.29 | 74.36 | 3.68 | 26.35–0.2 | 71.37 | <0.001 |
| [EA] | 0.40 | 0.95–0 | 29.05 | 0.48 | 0.99–0 | 23.09 | <0.001 |
| [LD] | 15.90 | 74.81–1.07 | 78.41 | 34.20 | 189.84–1.07 | 68.83 | <0.001 |
| [PA] | 0.63 | 1.22–0.56 | 16.24 | 0.56 | 1–0.54 | 3.95 | <0.001 |
| [PT] | 0.23 | 0.25–0.06 | 9.08 | 0.22 | 0.26–0.05 | 14.01 | <0.001 |
| Colour descriptors | |||||||
| L * | 41.28 | 92.65–23.58 | 26.20 | 38.65 | 78.46–13.31 | 19.62 | <0.001 |
| a * | 19.29 | 45.69–(−19.61) | 69.84 | 30.26 | 54.54–(−18.45) | 23.75 | <0.001 |
| b * | 22.99 | 82.33–2.26 | 66.38 | 22.31 | 83.62–(−9.48) | 51.54 | 0.284 |
| Chroma | 33.86 | 82.34–2.55 | 38.28 | 38.67 | 83.62–8.94 | 26.10 | <0.001 |
| Hue angle | 32.11 | 89.77–(−89.07) | 107.50 | 32.44 | 89.79–(−89.63) | 48.77 | 0.756 |
| Species. | Country Regions * |
|---|---|
| C. annuum | Brazil (1), Bulgaria (1), France (1), India (1), Japan (1), Kosovo (1), Mauritius Island (1), Nepal (1), Pakistan (1), Romania (1), Serbia (1), Madagascar (2), South Corea (2), Ukraine (2), Spain (3), Yemen (3), Canary Island (4), Vietnam (4), Mexico (9), Turkey (11), USA (11), Hungary (13), Italy (65), SB (40) |
| C. annuum var. glabriusculum | SB (2) |
| C. baccatum var. baccatum | Perù (2), Bolivia (1), SB (2) |
| C. baccatum var. pendulum | Chile (1), Cuba (1), Ethiopia (1), Paraguay (1), Bolivia (4), Brazil (5), Perù (10), SB (10) |
| C. chacoense | Argentina (1), Bolivia (4), SB (2) |
| C. chinense | Bali (1), Bangladesh (1), Burkina Faso (1), Central African Republic (1), Chile (1), Costa Rica (1), Guatemala (1), Vanuatu Islands (1), Barbados Islands (1), USA (1), Yemen (1), Bolivia (2), Maldives (2), Jamaica (2), Mexico (3), Trinidad (3), Caribbean (4), India (5), Perù (7), Brazil (8), SB (10) |
| C. eximium | India (1) |
| C. frutescens | Ecuador (1), Philippines (1), Hungary (1), kenya (1), Perù (1), Portugal (1), USA (1), Brazil (2), SB (3) |
| C. pubescens | Bolivia (1), Cuba (1), Ecuador (1), SB (2), Guatemala (2), Perù (3) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripodi, P.; Greco, B. Large Scale Phenotyping Provides Insight into the Diversity of Vegetative and Reproductive Organs in a Wide Collection of Wild and Domesticated Peppers (Capsicum spp.). Plants 2018, 7, 103. https://doi.org/10.3390/plants7040103
Tripodi P, Greco B. Large Scale Phenotyping Provides Insight into the Diversity of Vegetative and Reproductive Organs in a Wide Collection of Wild and Domesticated Peppers (Capsicum spp.). Plants. 2018; 7(4):103. https://doi.org/10.3390/plants7040103
Chicago/Turabian StyleTripodi, Pasquale, and Barbara Greco. 2018. "Large Scale Phenotyping Provides Insight into the Diversity of Vegetative and Reproductive Organs in a Wide Collection of Wild and Domesticated Peppers (Capsicum spp.)" Plants 7, no. 4: 103. https://doi.org/10.3390/plants7040103
APA StyleTripodi, P., & Greco, B. (2018). Large Scale Phenotyping Provides Insight into the Diversity of Vegetative and Reproductive Organs in a Wide Collection of Wild and Domesticated Peppers (Capsicum spp.). Plants, 7(4), 103. https://doi.org/10.3390/plants7040103