Plant Organelle Genome Replication
Abstract
:1. Introduction
1.1. Discovery of Mitochondria and Chloroplasts
1.2. Evolutionary Origins of Each Organelle
2. Organelle Genomes and Structure
2.1. Genome Size
2.2. Genome Structure and Content
3. Organelle DNA Replication
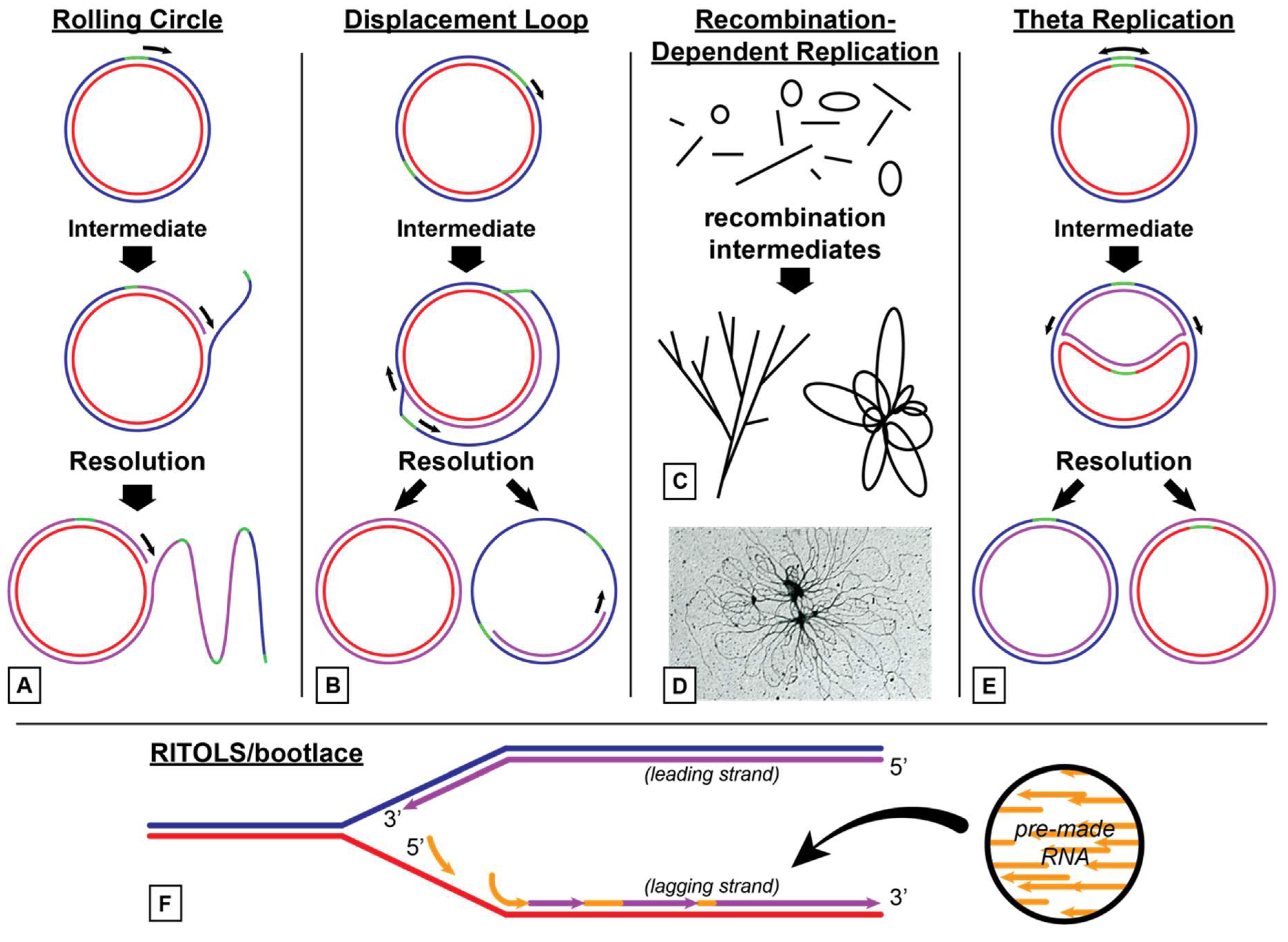
3.1. Models for Organelle DNA Replication
3.1.1. Animal Mitochondria
3.1.2. Plant Mitochondria
3.1.3. Plant Chloroplasts
3.2. Similarity to T7 Bacteriophage
4. Proteins Involved in Plant Mitochondrial and Chloroplast DNA Replication
4.1. Organelle DNA Replication Proteins
4.2. DNA Polymerases
4.3. DNA Unwinding
4.4. Organelle DNA Gyrases
4.5. Priming of DNA Synthesis
4.6. Primer Removal
4.7. Single-Stranded DNA Binding Proteins
4.8. DNA Recombination
4.9. DNA Ligation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hooke, R. Micrographia: Or Some Physiological Descriptions of Minute Bodies Made by Magnifying Glasses with Observations and Inquiries Thereupon; Facsimile Edition; Science Heritage, Ltd.: Lincolnwood, IL, USA, 1987; 323p. [Google Scholar]
- Harris, H. The Birth of the Cell; Yale University Press: New Haven, CT, USA, 1999; p. 212. [Google Scholar]
- Ernster, L.; Schatz, G. Mitochondria—A Historical Review. J. Cell Biol. 1981, 91, S227–S255. [Google Scholar] [CrossRef] [PubMed]
- Schimper, A.F.W. Ueber die Entwickelung der Chlorophyllkörner und Farbkörper. Botanische Zeitung 1883, 41, 105–160. [Google Scholar]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Kutschera, U.; Niklas, K.J. Endosymbiosis, cell evolution, and speciation. Theory Biosci. 2005, 124, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, J.L.; Schmidt, G.W. Pervasive migration of organellar DNA to the nucleus in plants. J. Mol. Evol. 1995, 41, 397–406. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Iborra, F.J.; Kimura, H.; Cook, P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004, 2, 9. [Google Scholar] [CrossRef]
- Kukat, C.; Wurm, C.A.; Spahr, H.; Falkenberg, M.; Larsson, N.G.; Jakobs, S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. USA 2011, 108, 13534–13539. [Google Scholar] [CrossRef] [Green Version]
- Montier, L.L.C.; Deng, J.J.; Bai, Y.D. Number matters: Control of mammalian mitochondrial DNA copy number. J. Genet. Genom. 2009, 36, 125–131. [Google Scholar] [CrossRef]
- Fauron, C.; Allen, J.; Clifton, S.; Newton, K. Plant Mitochondrial Genomes. In Molecular Biology and Biotechnology of Plant Organelles: Chloroplasts and Mitochondria; Daniell, H., Chase, C., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 151–177. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Kumar, R.A.; Bendich, A.J. The amount and integrity of mt DNA in maize decline with development. Planta 2013, 237, 603–617. [Google Scholar] [CrossRef]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Borner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010, 64, 948–959. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17. [Google Scholar] [CrossRef]
- Palmer, J.D. Comparative Organization of Chloroplast Genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Kolodner, R.; Tewari, K.K. Inverted Repeats in Chloroplast DNA from Higher-Plants. Proc. Natl. Acad. Sci. USA 1979, 76, 41–45. [Google Scholar] [CrossRef]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Gray, M.W. The Bacterial Ancestry of Plastids and Mitochondria. BioScience 1983, 33, 693–699. [Google Scholar] [CrossRef]
- Zoschke, R.; Liere, K.; Borner, T. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 2007, 50, 710–722. [Google Scholar] [CrossRef]
- Zheng, Q.; Oldenburg, D.J.; Bendich, A.J. Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J. Exp. Bot. 2011, 62, 2715–2730. [Google Scholar] [CrossRef] [Green Version]
- Udy, D.B.; Belcher, S.; Williams-Carrier, R.; Gualberto, J.M.; Barkan, A. Effects of Reduced Chloroplast Gene Copy Number on Chloroplast Gene Expression in Maize. Plant Physiol. 2012, 160, 1420–1431. [Google Scholar] [CrossRef] [Green Version]
- Shaver, J.M.; Oldenburg, D.J.; Bendich, A.J. Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 2006, 224, 72–82. [Google Scholar] [CrossRef]
- Rowan, B.A.; Oldenburg, D.J.; Bendich, A.J. The demise of chloroplast DNA in Arabidopsis. Curr. Genet. 2004, 46, 176–181. [Google Scholar] [CrossRef]
- Rowan, B.A.; Oldenburg, D.J.; Bendich, A.J. A multiple-method approach reveals a declining amount of chloroplast DNA during development in Arabidopsis. BMC Plant Biol. 2009, 9. [Google Scholar] [CrossRef]
- Clayton, D.A. Replication of animal mitochondrial DNA. Cell 1982, 28, 693–705. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; Debruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Pett, W. Animal Mitochondrial DNA as We Do Not Know It: Mt-Genome Organization and Evolution in Nonbilaterian Lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef]
- Shadel, G.S.; Clayton, D.A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997, 66, 409–435. [Google Scholar] [CrossRef]
- Okimoto, R.; Macfarlane, J.L.; Clary, D.O.; Wolstenholme, D.R. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 1992, 130, 471–498. [Google Scholar]
- Hoffmann, R.J.; Boore, J.L.; Brown, W.M. A Novel Mitochondrial Genome Organization for the Blue Mussel, Mytilus-Edulis. Genetics 1992, 131, 397–412. [Google Scholar] [PubMed]
- Beagley, C.T.; Macfarlane, J.L.; PontKingdon, G.A.; Okimoto, R.; Okada, N.A.; Wolstenholme, D.R. Mitochondrial genomes of anthozoa (Cnidaria). Prog. Cell Res. 1995, 5, 149–153. [Google Scholar]
- Oda, K.; Kohchi, T.; Ohyama, K. Mitochondrial-DNA of Marchantia-Polymorpha as a Single Circular Form with No Incorporation of Foreign DNA. Biosci. Biotechnol. Biochem. 1992, 56, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Nielsen, B.L.; Borner, T. The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997, 2, 477–483. [Google Scholar] [CrossRef]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Cupp, J.D.; Nielsen, B.L. Minireview: DNA replication in plant mitochondria. Mitochondrion 2014, 19, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, W.; Brennicke, A. The Plant Mitochondrial Genome—Physical Structure, Information-Content, Rna Editing, and Gene Migration to the Nucleus. Annu. Rev. Plant Phys. 1994, 45, 61–78. [Google Scholar] [CrossRef]
- Palmer, J.D.; Adams, K.L.; Cho, Y.R.; Parkinson, C.L.; Qiu, Y.L.; Song, K.M. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 2000, 97, 6960–6966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morley, S.A.; Nielsen, B.L. Plant mitochondrial DNA. Front. Biosci-Landmrk 2017, 22, 1023–1032. [Google Scholar] [CrossRef] [Green Version]
- Burger, G.; Gray, M.W.; Lang, B.F. Mitochondrial genomes: Anything goes. Trends Genet. 2003, 19, 709–716. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Herbon, L.A. Plant Mitochondrial-DNA Evolves Rapidly in Structure, but Slowly in Sequence. J. Mol. Evol. 1988, 28, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.C. Plant Mitochondrial Genome Evolution Can Be Explained by DNA Repair Mechanisms. Genome Biol. Evol. 2013, 5, 1079–1086. [Google Scholar] [CrossRef]
- Parsons, T.J.; Muniec, D.S.; Sullivan, K.; Woodyatt, N.; AllistonGreiner, R.; Wilson, M.R.; Berry, D.L.; Holland, K.A.; Weedn, V.W.; Gill, P.; et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat. Genet. 1997, 15, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M. The chloroplast genome. Essays Biochem. 1995, 30, 49–57. [Google Scholar]
- Palmer, J.D.; Stein, D.B. Conservation of Chloroplast Genome Structure among Vascular Plants. Curr. Genet. 1986, 10, 823–833. [Google Scholar] [CrossRef]
- Dalmon, J.; Loiseaux, S.; Bazetoux, S. Heterogeneity of plastid DNA of two species of brown algae. Plant Sci. Lett. 1983, 29, 243–253. [Google Scholar] [CrossRef]
- Ciesielski, G.L.; Oliveira, M.T.; Kaguni, L.S. Animal Mitochondrial DNA Replication. Enzymes 2016, 39, 255–292. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.C.; Joers, P.; Willcox, S.; Griffith, J.D.; Jacobs, H.T.; Hyman, B.C. A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in Caenorhabditis Elegans. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Fish, J.; Raule, N.; Attardi, G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 2004, 306, 2098–2101. [Google Scholar] [CrossRef]
- Holt, I.J.; Lorimer, H.E.; Jacobs, H.T. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 2000, 100, 515–524. [Google Scholar] [CrossRef]
- Korhonen, J.A.; Pham, X.H.; Pellegrini, M.; Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004, 23, 2423–2429. [Google Scholar] [CrossRef]
- Dehaas, J.M.; Hille, J.; Kors, F.; Vandermeer, B.; Kool, A.J.; Folkerts, O.; Nijkamp, H.J.J. 2 Potential Petunia-Hybrida Mitochondrial-DNA Replication Origins Show Structural and Invitro Functional Homology with the Animal Mitochondrial-DNA Heavy and Light Strand Replication Origins. Curr. Genet. 1991, 20, 503–513. [Google Scholar] [CrossRef]
- Backert, S.; Dorfel, P.; Lurz, R.; Borner, T. Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L). Mol. Cell Biol. 1996, 16, 6285–6294. [Google Scholar] [CrossRef]
- Backert, S.; Borner, T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 2000, 37, 304–314. [Google Scholar] [CrossRef]
- Lockshon, D.; Zweifel, S.G.; Freeman-Cook, L.L.; Lorimer, H.E.; Brewer, B.J.; Fangman, W.L. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 1995, 81, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Heinhorst, S.; Cannon, G.C. DNA-Replication in Chloroplasts. J. Cell Sci. 1993, 104, 1–9. [Google Scholar]
- Kolodner, R.; Tewari, K.K. Presence of displacement loops in the covalently closed circular chloroplast deoxyribonucleic acid from higher plants. J. Biol. Chem. 1975, 250, 8840–8847. [Google Scholar]
- Kunnimalaiyaan, M.; Nielsen, B.L. Chloroplast DNA replication: Mechanism, enzymes and replication origins. J. Plant Biochem. Biot. 1997, 6, 1–7. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Misumi, O.; Odahara, M.; Ishibashi, K.; Hirono, M.; Hidaka, K.; Endo, M.; Sugiyama, H.; Iwasaki, H.; Kuroiwa, T.; et al. Holliday junction resolvases mediate chloroplast nucleoid segregation. Science 2017, 356, 631–634. [Google Scholar] [CrossRef]
- Chiu, W.L.; Sears, B.B. Electron-Microscopic Localization of Replication Origins in Oenothera Chloroplast DNA. Mol. Gen. Genet. 1992, 232, 33–39. [Google Scholar] [CrossRef]
- Waddell, J.; Wang, X.M.; Wu, M. Electron-Microscopic Localization of the Chloroplast DNA Replicative Origins in Chlamydomonas-Reinhardii. Nucleic Acids Res. 1984, 12, 3843–3856. [Google Scholar] [CrossRef]
- Ravelchapuis, P.; Heizmann, P.; Nigon, V. Electron-Microscopic Localization of the Replication Origin of Euglena-Gracilis Chloroplast DNA. Nature 1982, 300, 78–81. [Google Scholar] [CrossRef]
- Lee, S.J.; Richardson, C.C. Choreography of bacteriophage T7 DNA replication. Curr. Opin. Chem. Biol. 2011, 15, 580–586. [Google Scholar] [CrossRef] [Green Version]
- Sato, N.; Terasawa, K.; Miyajima, K.; Kabeya, Y. Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 2003, 232, 217–262. [Google Scholar] [CrossRef]
- Elo, A.; Lyznik, A.; Gonzalez, D.O.; Kachman, S.D.; Mackenzie, S.A. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 2003, 15, 1619–1631. [Google Scholar] [CrossRef]
- Ono, Y.; Sakai, A.; Takechi, K.; Takio, S.; Takusagawa, M.; Takano, H. NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007, 48, 1679–1692. [Google Scholar] [CrossRef]
- Carrie, C.; Kuhn, K.; Murcha, M.W.; Duncan, O.; Small, I.D.; O’Toole, N.; Whelan, J. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009, 57, 1128–1139. [Google Scholar] [CrossRef]
- Diray-Arce, J.; Liu, B.; Cupp, J.D.; Hunt, T.; Nielsen, B.L. The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 2013, 13. [Google Scholar] [CrossRef]
- Shutt, T.E.; Gray, M.W. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006, 62, 588–599. [Google Scholar] [CrossRef]
- Duxin, J.P.; Dao, B.; Martinsson, P.; Rajala, N.; Guittat, L.; Campbell, J.L.; Spelbrink, J.N.; Stewart, S.A. Human Dna2 Is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009, 29, 4274–4282. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, M.A.; Guo, Z.G.; Lu, H.M.; Qian, L.M.; Dai, H.F.; Qiu, J.Z.; Yakubovskaya, E.; Bogenhagen, D.F.; Demple, B.; et al. Human DNA2 Is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 2008, 32, 325–336. [Google Scholar] [CrossRef]
- Liere, K.; Weihe, A.; Borner, T. The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 2011, 168, 1345–1360. [Google Scholar] [CrossRef]
- Carrie, C.; Small, I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta 2013, 1833, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Hedtke, B.; Borner, T.; Weihe, A. One RNA polymerase serving two genomes. EMBO Rep. 2000, 1, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Hedtke, B.; Borner, T.; Weihe, A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 1997, 277, 809–811. [Google Scholar] [CrossRef]
- Yang, Z.; Hou, Q.C.; Cheng, L.L.; Xu, W.; Hong, Y.T.; Li, S.; Sun, Q.W. RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 2017, 29, 2478–2497. [Google Scholar] [CrossRef] [Green Version]
- Edmondson, A.C.; Song, D.Q.; Alvarez, L.A.; Wall, M.K.; Almond, D.; McClellan, D.A.; Maxwell, A.; Nielsen, B.L. Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol. Genet. Genom. 2005, 273, 115–122. [Google Scholar] [CrossRef]
- Zaegel, V.; Guermann, B.; Le Ret, M.; Andres, C.; Meyer, D.; Erhardt, M.; Canaday, J.; Gualberto, J.M.; Imbault, P. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 2006, 18, 3548–3563. [Google Scholar] [CrossRef]
- Krause, K.; Kilbienski, I.; Mulisch, M.; Rodiger, A.; Schafer, A.; Krupinska, K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005, 579, 3707–3712. [Google Scholar] [CrossRef]
- Cappadocia, L.; Marechal, A.; Parent, J.S.; Lepage, E.; Sygusch, J.; Brisson, N. Crystal Structures of DNA-Whirly Complexes and Their Role in Arabidopsis Organelle Genome Repair. Plant Cell 2010, 22, 1849–1867. [Google Scholar] [CrossRef] [Green Version]
- Marechal, A.; Parent, J.S.; Veronneau-Lafortune, F.; Joyeux, A.; Lang, B.F.; Brisson, N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 14693–14698. [Google Scholar] [CrossRef]
- Shedge, V.; Arrieta-Montiel, M.; Christensen, A.C.; Mackenzie, S.A. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 2007, 19, 1251–1264. [Google Scholar] [CrossRef]
- Khazi, F.R.; Edmondson, A.C.; Nielsen, B.L. An Arabidopsis homologue of bacterial RecA that complements an E-coli recA deletion is targeted to plant mitochondria. Mol. Genet. Genom. 2003, 269, 454–463. [Google Scholar] [CrossRef]
- Xu, Y.Z.; Arrieta-Montiel, M.P.; Virdi, K.S.; de Paula, W.B.M.; Widhalm, J.R.; Basset, G.J.; Davila, J.I.; Elthon, T.E.; Elowsky, C.G.; Sato, S.J.; et al. MutS HOMOLOG1 Is a Nucleoid Protein That Alters Mitochondrial and Plastid Properties and Plant Response to High Light. Plant Cell 2011, 23, 3428–3441. [Google Scholar] [CrossRef] [Green Version]
- Wall, M.K.; Mitchenall, L.A.; Maxwell, A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 2004, 101, 7821–7826. [Google Scholar] [CrossRef]
- Morley, S.A.; Nielsen, B.L. Chloroplast DNA Copy Number Changes during Plant Development in Organelle DNA Polymerase Mutants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Parent, J.S.; Lepage, E.; Brisson, N. Divergent Roles for the Two PolI-Like Organelle DNA Polymerases of Arabidopsis. Plant Physiol. 2011, 156, 254–262. [Google Scholar] [CrossRef] [Green Version]
- Trasvina-Arenas, C.H.; Baruch-Torres, N.; Cordoba-Andrade, F.J.; Ayala-Garcia, V.M.; Garcia-Medel, P.L.; Diaz-Quezada, C.; Peralta-Castro, A.; Ordaz-Ortiz, J.J.; Brieba, L.G. Identification of a unique insertion in plant organellar DNA polymerases responsible for 5‘-dRP lyase and strand-displacement activities: Implications for Base Excision Repair. DNA Repair. 2018, 65, 1–10. [Google Scholar] [CrossRef]
- Ayala-Garcia, V.M.; Baruch-Torres, N.; Garcia-Medel, P.; Brieba, L.G. Plant organellar DNA polymerases paralogs exhibit dissimilar nucleotide incorporation fidelity. FEBS J. 2018. [Google Scholar] [CrossRef]
- Moriyama, T.; Sato, N. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Eun, H.-M. 6—DNA Polymerases. In Enzymology Primer for Recombinant DNA Technology; Eun, H.-M., Ed.; Academic Press: San Diego, CA, USA, 1996; pp. 345–489. [Google Scholar]
- Bedford, E.; Tabor, S.; Richardson, C.C. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. USA 1997, 94, 479–484. [Google Scholar] [CrossRef]
- McCulloch, S.D.; Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008, 18, 148–161. [Google Scholar] [CrossRef] [Green Version]
- Baruch-Torres, N.; Brieba, L.G. Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 2017, 45, 10751–10763. [Google Scholar] [CrossRef] [Green Version]
- Peralta-Castro, A.; Baruch-Torres, N.; Brieba, L.G. Plant organellar DNA primase-helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence. Nucleic Acids Res. 2017, 45, 10764–10774. [Google Scholar] [CrossRef] [Green Version]
- Spelbrink, J.N.; Li, F.Y.; Tiranti, V.; Nikali, K.; Yuan, Q.P.; Tariq, M.; Wanrooij, S.; Garrido, N.; Comi, G.; Morandi, L.; et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001, 29, 223–231. [Google Scholar] [CrossRef]
- Korhonen, J.A.; Gaspari, M.; Falkenberg, M. TWINKLE has 5′-> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003, 278, 48627–48632. [Google Scholar] [CrossRef]
- Levikova, M.; Cejka, P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015, 43, 7888–7897. [Google Scholar] [CrossRef]
- Li, Z.; Liu, B.; Jin, W.; Wu, X.; Zhou, M.; Liu, V.Z.; Goel, A.; Shen, Z.; Zheng, L.; Shen, B. hDNA2 nuclease/helicase promotes centromeric DNA replication and genome stability. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Jia, N.; Liu, X.M.; Gao, H.B. A DNA2 Homolog Is Required for DNA Damage Repair, Cell Cycle Regulation, and Meristem Maintenance in Plants. Plant Physiol. 2016, 171, 318–333. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.S.; Lee, S.S.; Kim, K.D.; Hwang, I.; Lim, J.-S.; Park, Y.-I.; Pai, H.-S. DNA gyrase is involved in chloroplast nucleoid partitioning. Plant Cell 2004, 16, 2665–2682. [Google Scholar] [CrossRef]
- Fuste, J.M.; Wanrooij, S.; Jemt, E.; Granycome, C.E.; Cluett, T.J.; Shi, Y.H.; Atanassova, N.; Holt, I.J.; Gustafsson, C.M.; Falkenberg, M. Mitochondrial RNA Polymerase Is Needed for Activation of the Origin of Light-Strand DNA Replication. Mol. Cell 2010, 37, 67–78. [Google Scholar] [CrossRef]
- Ramachandran, A.; Nandakumar, D.; Deshpande, A.P.; Lucas, T.P.; R-Bhojappa, R.; Tang, G.Q.; Raney, K.; Yin, Y.W.; Patel, S.S. The Yeast Mitochondrial RNA Polymerase and Transcription Factor Complex Catalyzes Efficient Priming of DNA Synthesis on Single-stranded DNA. J. Biol. Chem. 2016, 291, 16828–16839. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.R. Updating Our View of Organelle Genome Nucleotide Landscape. Front. Genet. 2012, 3, 175. [Google Scholar] [CrossRef] [Green Version]
- Larson, M.A.; Bressani, R.; Sayood, K.; Corn, J.E.; Berger, J.M.; Griep, M.A.; Hinrichs, S.H. Hyperthermophilic Aquifex aeolicus initiates primer synthesis on a limited set of trinucleotides comprised of cytosines and guanines. Nucleic Acids Res. 2008, 36, 5260–5269. [Google Scholar] [CrossRef]
- Arnold, J.J.; Smidansky, E.D.; Moustafa, I.M.; Cameron, C.E. Human mitochondrial RNA polymerase: Structure-function, mechanism and inhibition. Biochim. Biophys. Acta 2012, 1819, 948–960. [Google Scholar] [CrossRef]
- Hess, W.R.; Borner, T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999, 190, 1–59. [Google Scholar] [CrossRef]
- Yin, C.; Richter, U.; Borner, T.; Weihe, A. Evolution of plant phage-type RNA polymerases: The genome of the basal angiosperm Nuphar advena encodes two mitochondrial and one plastid phage-type RNA polymerases. BMC Evol. Biol. 2010, 10, 379. [Google Scholar] [CrossRef]
- Baba, K.; Schmidt, J.; Espinosa-Ruiz, A.; Villarejo, A.; Shiina, T.; Gardestrom, P.; Sane, A.P.; Bhalerao, R.P. Organellar gene transcription and early seedling development are affected in the rpoT; 2 mutant of Arabidopsis. Plant J. 2004, 38, 38–48. [Google Scholar] [CrossRef]
- Serino, G.; Maliga, P. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 1998, 117, 1165–1170. [Google Scholar] [CrossRef]
- Nielsen, B.L.; Rajasekhar, V.K.; Tewari, K.K. Pea Chloroplast DNA Primase—Characterization and Role in Initiation of Replication. Plant Mol. Biol. 1991, 16, 1019–1034. [Google Scholar] [CrossRef]
- Morley, S.A.; Peralta-Castro, A.; Brieba, L.G.; Miller, J.; Ong, K.L.; Ridge, P.G.; Oliphant, A.; Aldous, S.; Nielsen, B.L. Arabidopsis thaliana organelles mimic the T7 phage DNA replisome with specific interactions between Twinkle protein and DNA polymerases Pol1A and Pol1B. BMC Plant Biol. 2019, 19, 241. [Google Scholar] [CrossRef]
- Desveaux, D.; Marechal, A.; Brisson, N. Whirly transcription factors: Defense gene regulation and beyond. Trends Plant Sci. 2005, 10, 95–102. [Google Scholar] [CrossRef]
- Redei, G.P. Extrachromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 1973, 18, 149–162. [Google Scholar] [CrossRef]
- Martinezzapater, J.M.; Gil, P.; Capel, J.; Somerville, C.R. Mutations at the Arabidopsis Chm locus promote rearrangements of the mitochondrial genome. Plant Cell 1992, 4, 889–899. [Google Scholar]
- Abdelnoor, R.V.; Yule, R.; Elo, A.; Christensen, A.C.; Meyer-Gauen, G.; Mackenzie, S.A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 2003, 100, 5968–5973. [Google Scholar] [CrossRef] [Green Version]
- Reenan, R.A.G.; Kolodner, R.D. Characterization of insertion mutations in the Saccharomyces cerevisiae Msh1 and Msh2 genes—Evidence for separate mitochondrial and nuclear Functions. Genetics 1992, 132, 975–985. [Google Scholar]
- Abdelnoor, R.V.; Christensen, A.C.; Mohammed, S.; Munoz-Castillo, B.; Moriyama, H.; Mackenzie, S.A. Mitochondrial genome dynamics in plants and animals: Convergent gene fusions of a MutS homologue. J. Mol. Evol. 2006, 63, 165–173. [Google Scholar] [CrossRef]
- Davila, J.I.; Arrieta-Montiel, M.P.; Wamboldt, Y.; Cao, J.; Hagmann, J.; Shedge, V.; Xu, Y.-Z.; Weigel, D.; Mackenzie, S.A. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011, 9, 64. [Google Scholar] [CrossRef]
- Odahara, M.; Kishita, Y.; Sekine, Y. MSH1 maintains organelle genome stability and genetically interacts with RECA and RECG in the moss Physcomitrella patens. Plant J. 2017, 91, 455–465. [Google Scholar] [CrossRef]
- Fukui, K.; Harada, A.; Wakamatsu, T.; Minobe, A.; Ohshita, K.; Ashiuchi, M.; Yano, T. The GIY-YIG endonuclease domain of Arabidopsis MutS homolog 1 specifically binds to branched DNA structures. FEBS Lett. 2018, 592, 4066–4077. [Google Scholar] [CrossRef]
- Sunderland, P.A.; West, C.E.; Waterworth, W.M.; Bray, C.M. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 2006, 47, 356–367. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9. [Google Scholar] [CrossRef]


| Function | Protein Name | TAIR | Homology | Localization * | Ref. |
|---|---|---|---|---|---|
| DNA polymerase | Pol1A or Pol gamma 2 | At1g50840 | Bacterial | M, P | [69,70,71] |
| Pol1B or Pol gamma 2 | At3g20540 | Bacterial | M, P | [69,70,71] | |
| Helicase | Twinkle | At1g30680 | Phage | M, P | [72,73] |
| DNA2 | At1g08840 | Mammalian | ? | [74,75] | |
| Priming | Twinkle | At1g30680 | Phage | M, P | [72,73] |
| RNA polymerase | [76,77,78,79] | ||||
| RpoT1 | At1g68990 | Phage | M | ||
| RpoT2 | At5g15700 | Phage | M, P | ||
| RpoT3 | At2g24120 | Phage | P | ||
| RpoA | AtCg00740 | Bacterial | P | ||
| RpoB | AtCg00190 | Bacterial | P | ||
| RpoC1 | AtCg00180 | Bacterial | P | ||
| RpoC2 | AtCg00170 | Bacterial | P | ||
| Primer Removal | RNaseH | ||||
| AtRNH1B | At5g51080 | [80] | |||
| AtRNH1C | At1g24090 | Bacterial | M, P | [80] | |
| EXO1 | ? | Bacterial | P | [68] | |
| EXO2 | ? | [68] | |||
| ssDNA binding, recombination monitoring | SSB1 | At4g11060 | Bacterial | M, P | [44,81] |
| SSB2 | At3g18580 | Bacterial | M | [44] | |
| OSB1 | At3g18580 | Bacterial-like, but unique to plants | M | [82] | |
| OSB2 | At4g20010 | Unique to plants | P | [44] | |
| OSB3 | At5g44785 | Unique to plants | M, P | [77,82] | |
| OSB4 | At1g31010 | Unique to plants | M | [82] | |
| WHY1 | At1g14410 | Unique to plants | P | [83,84,85] | |
| WHY2 | At1g71260 | Unique to plants | M | [83,84,85] | |
| WHY3 | At2g02740 | Unique to plants | P | [83,84,85] | |
| ODB1 | At1g71310 | M, N? | |||
| ODB2 | At5g47870 | P, N? | |||
| Recombination | RecA1 | At1g79050 | Bacterial | P | [86] |
| RecA2 | At2g19490 | Bacterial | M, P | [86] | |
| RecA3 | At3g10140 | Bacterial | M | [86,87] | |
| MSH1 | At3g24320 | Bacterial | M, P | [88] | |
| Topoisomerase | Topoisomerase I | At4g31210 | Bacterial | M, P | [77] |
| DNA Gyrase A | At3g10690 | Bacterial | M, P | [89] | |
| DNA Gyrase B1 | At3g10270 | Bacterial | P | [89] | |
| DNA Gyrase B2 | At5g04130 | Bacterial | M | [89] | |
| DNA Gyrase B3 | At5g04110 | Eukaryotic | N | [89] | |
| Ligation | LIG1 | At1g08130 | Bacterial | M, N | [44] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morley, S.A.; Ahmad, N.; Nielsen, B.L. Plant Organelle Genome Replication. Plants 2019, 8, 358. https://doi.org/10.3390/plants8100358
Morley SA, Ahmad N, Nielsen BL. Plant Organelle Genome Replication. Plants. 2019; 8(10):358. https://doi.org/10.3390/plants8100358
Chicago/Turabian StyleMorley, Stewart A., Niaz Ahmad, and Brent L. Nielsen. 2019. "Plant Organelle Genome Replication" Plants 8, no. 10: 358. https://doi.org/10.3390/plants8100358
APA StyleMorley, S. A., Ahmad, N., & Nielsen, B. L. (2019). Plant Organelle Genome Replication. Plants, 8(10), 358. https://doi.org/10.3390/plants8100358







