The Recovery from Sulfur Starvation Is Independent from the mRNA Degradation Initiation Enzyme PARN in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. The Sulfur-Responsive Transcripts SULTR1;1, SULTR1;2, SDI1, SHM7 and GGCT Contain ARE Sites
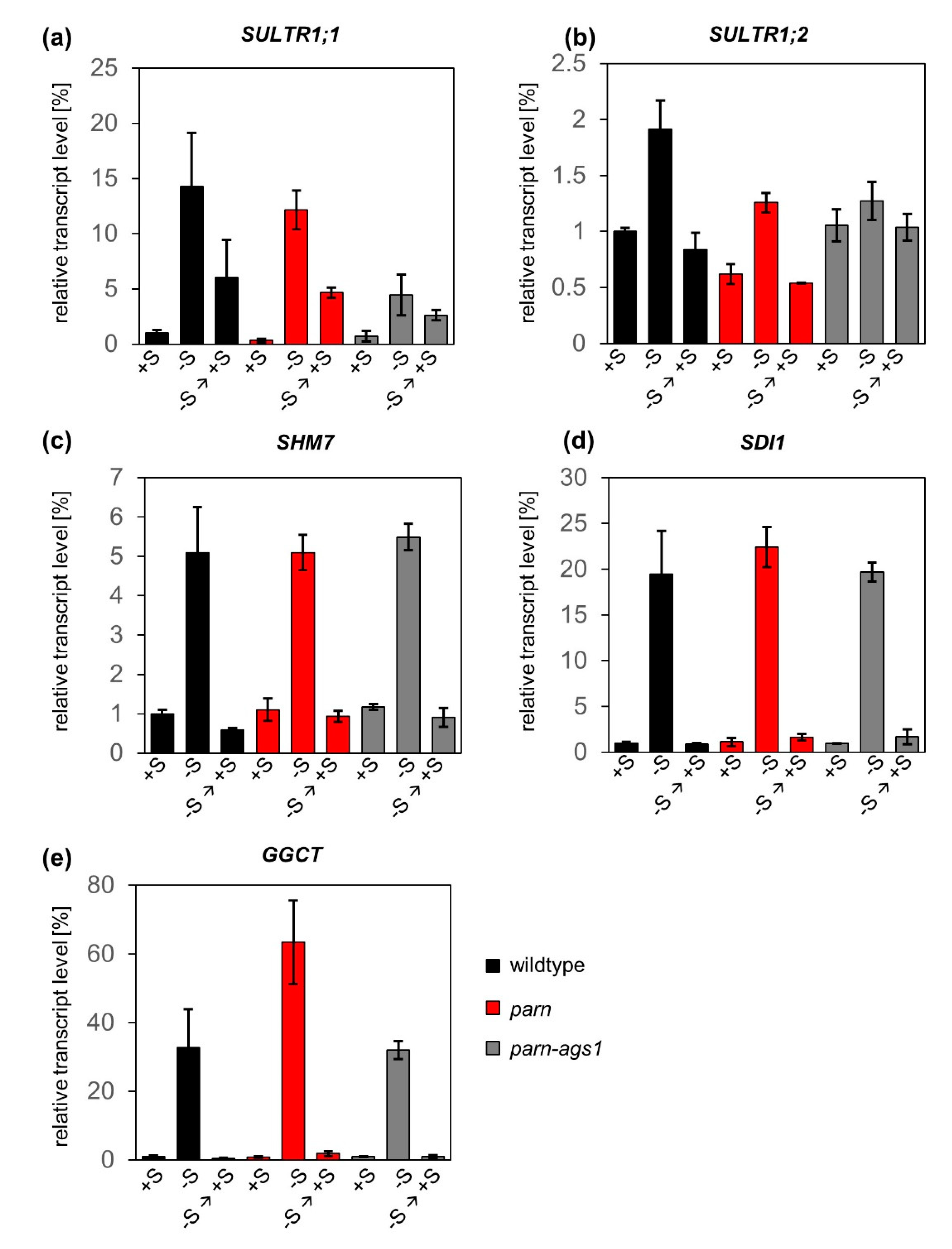
2.2. The Degradation of Sulfur Metabolism-Related Transcripts Is Independent of AtPARN
2.3. PARN Accumulates in Cytoplasmic Speckles
2.4. PARN is Encoded for by Two Alternative Splice Variants Predicted to Localize to Different Cellular Compartments
3. Discussion
3.1. The Degradation of Sulfur Starvation-Induced Genes during the Recovery from Starvation is Independent of AtPARN
3.2. The Presence of Two Alternative PARN Splice Variants Reconciles the Diverging Views on PARN Localization in the Literature
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Sulfur Resupply Assay
4.3. Genotyping by PCR
4.4. Quantifying Gene Expression by qRT-PCR
4.5. Immunodetection of PARN-GFP
4.6. Subcellular Localization
4.7. Identification and Functional Annotation of Genes with mRNA Destabilizing Motifs
4.8. Prediction of Subcellular Protein Localization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Allele | TAIR Identifier | Sequence |
|---|---|---|
| ACTIN7 | AT5G09810 | CAACCGGTATTGTGCTCGATTC |
| GAGTGAGTCTGTGAGATCCCG | ||
| AGS1 | AT2G17580 | CTAGCAAATTCGACAGCTTTGC |
| ATTTTAGAGGTTATTCTCCAATATGG | ||
| AGS1 mutated | AT2G17580 | ATTTTAGAGGTTATTCTCCAATATGA |
| AtPARN | AT1G55870 | CTGATTCAGATTCCGACAAGGA |
| CTTTGCCTCCTTCTGTGAAAAG | ||
| AtPARN mutated | AT1G55870 | GTATACTGATTCAGATTCCGACAAA |
| GFP | N.A. | GCGGATCTTGAAGTTGGCC |
| GFP | N.A. | CGACGGCAACTACAAGACC |
| Primer | Sequence |
|---|---|
| I | CCCTTTCGTTGGGATTCTCG |
| GTATGCAGGTGTTGAAATCAAAC | |
| II | CACGAATTTCTCAGCTGTTGAAG |
| TGCTTCAGAGAAAAAGCTGATCG | |
| III | CTACCCGTTAGTCTCTCTTTC |
| GACGAGGAAATACAAAGAAATTGTG | |
| IV | GCAAAACCTAAAAATGGTCGTTTG |
| CGAGAATCCCAACGAAAGGG | |
| V | CCCTTTCGTTGGGATTCTCG |
| CATGAGCTGGTGGATCAAATG |
| Allele | TAIR Identifier | Sequence |
|---|---|---|
| GGCT | AT1G44790 | CCGGAGCTATTTGCTGGGGTG |
| GTCGTATTCACACTCTCTTCGTTCC | ||
| PP2A | AT1G69960 | CTTCTCGCTCCAGTAATGGGATCC |
| GCTTGGTCGACTATCGGAATGAGAG | ||
| SDI1 | AT5G48850 | CCCTTGACAATGTCCTCATCG |
| GCTTCTCCTTGATAGATCTGCC | ||
| SHM7 | AT1G36370 | CTATACAGCCTCGGGTTGTCATTG |
| AACTAACGTCATTACATACACATCTTG | ||
| SULTR1;1 | AT5G04590 | GTCCGGGACTATTAATCCC |
| CGTACCCCATGCTCAGCG | ||
| SULTR1;2 | AT1G78000 | GGATCCAGAGATGGCTACATGA |
| TCGATGTCCGTAACAGGTGAC | ||
| TIP41 | AT3G54000 | GATGAGGCACCAACTGTTCTTCGTG |
| CTGACTGATGGAGCTCGGGTCG |
References
- Kopriva, S.; Mugford, S.G.; Baraniecka, P.; Lee, B.R.; Matthewman, C.A.; Koprivova, A. Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front. Plant Sci. 2012, 3, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, T.L.; Baker, G.W.; Wilks, F.R.; Popov, V.A.; Mathur, J.; Benfey, P.N. Large cellular inclusions accumulate in Arabidopsis roots exposed to low-sulfur conditions. Plant Physiol. 2015, 168, 1573–1589. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.J.; Hawkesford, M.J.; McGrath, S.P. Sulphur assimilation and effects on yield and quality of wheat. J. Cereal. Sci 1999, 30, 1–17. [Google Scholar] [CrossRef]
- Ding, Y.; Zhou, X.; Zuo, L.; Wang, H.; Yu, D. Identification and functional characterization of the sulfate transporter gene GmSULTR1;2b in soybean. BMC Genom. 2016, 17, 373. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Sirko, A. Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Pol. 2008, 55, 457–471. [Google Scholar]
- Nikiforova, V.; Freitag, J.; Kempa, S.; Adamik, M.; Hesse, H.; Hoefgen, R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 2003, 33, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A. Metabolic changes sustain the plant life in low-sulfur environments. Curr. Opin. Plant Biol. 2017, 39, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, M.; Watanabe, M.; Morcuende, R.; Scheible, W.-R.; Hawkesford, M.J.; Hesse, H.; Hoefgen, R. Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Front. Plant Sci. 2015, 5, 805. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Howell, K.A.; Millar, A.H.; O’Toole, N.; Small, I.; Whelan, J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 2007, 19, 3418–3436. [Google Scholar] [CrossRef]
- Crisp, P.A.; Ganguly, D.; Smith, A.B.; Murray, K.D.; Estavillo, G.M.; Searle, I.R.; Ford, E.; Bogdanović, O.; Lister, R.; Borevitz, J.O.; et al. Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 2017, 29, 1836–1863. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, K.; Matsui, A.; Shinozaki, K.; Seki, M. RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Brosnan, C.A.; Rothnagel, J.A.; Carroll, B.J. RNA decay and RNA silencing in plants: Competition or collaboration? Front. Plant Sci. 2011, 2, 99. [Google Scholar] [CrossRef] [PubMed]
- Belostotsky, D.A.; Sieburth, L.E. Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 2009, 12, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Green, P.J. mRNA degradation machinery in plants. J. Plant Biol. 2009, 52, 114–124. [Google Scholar] [CrossRef]
- Reverdatto, S.V.; Dutko, J.A.; Chekanova, J.A.; Hamilton, D.A.; Belostotsky, D.A. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 2004, 10, 1200–1214. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, N.; Kitahata, N.; Seki, M.; Narusaka, Y.; Narusaka, M.; Kuromori, T.; Asami, T.; Shinozaki, K.; Hirayama, T. Analysis of ABA hypersensitive germination 2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005, 44, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Okamoto, M.; Narusaka, M.; Yasuda, M.; Nakashita, H.; Shinozaki, K.; Narusaka, Y.; Hirayama, T. ABA hypersensitive germination 2-1 causes the activation of both abscisic acid and salicylic acid responses in Arabidopsis. Plant Cell Physiol. 2009, 50, 2112–2122. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Murayama, M.; Asami, T.; Shinozaki, K.; Hirayama, T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004, 45, 1485–1499. [Google Scholar] [CrossRef]
- Hirayama, T. A unique system for regulating mitochondrial mRNA poly(A) status and stability in plants. Plant Singal. Behav. 2014, 9, e973809. [Google Scholar] [CrossRef]
- Hirayama, T.; Matsuura, T.; Ushiyama, S.; Narusaka, M.; Kurihara, Y.; Yasuda, M.; Ohtani, M.; Seki, M.; Demura, T.; Nakashita, H.; et al. A poly(A)-specific ribonuclease directly regulates the poly(A) status of mitochondrial mRNA in Arabidopsis. Nat. Commun. 2013, 4, 2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malcheska, F.; Ahmad, A.; Batool, S.; Muller, H.M.; Ludwig-Muller, J.; Kreuzwieser, J.; Randewig, D.; Hansch, R.; Mendel, R.R.; Hell, R.; et al. Drought-enhanced xylem sap sulfate closes stomata by affecting ALMT12 and guard cell ABA synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef] [PubMed]
- Batool, S.; Uslu, V.V.; Rajab, H.; Ahmad, N.; Waadt, R.; Geiger, D.; Malagoli, M.; Hedrich, R.; Rennenberg, H.; Herschbach, C.; et al. Sulfate is incorporated into cysteine to trigger ABA production and stomata closure. Plant Cell 2018, 30, 2973–2987. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.S.; Kennington, E.A.; Blackshear, P.J. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003, 23, 3798–3812. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Shyu, A.B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Gherzi, R.; Ong, S.E.; Chan, E.L.; Raijmakers, R.; Pruijn, G.J.; Stoecklin, G.; Moroni, C.; Mann, M.; Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 2001, 107, 451–464. [Google Scholar] [CrossRef]
- Aarabi, F.; Hubberten, H.-M.; Heyneke, E.; Watanabe, M.; Hoefgen, R. OAS Cluster Genes: A Tightly Co-regulated Network. In Molecular Physiology and Ecophysiology of Sulfur; De Kok, L.J., Hawkesford, M.J., Rennenberg, H., Saito, K., Schnug, E., Eds.; Springer: Cham, Germany, 2015; pp. 125–132. [Google Scholar] [CrossRef]
- Moreno, A.B.; Martinez de Alba, A.E.; Bardou, F.; Crespi, M.D.; Vaucheret, H.; Maizel, A.; Mallory, A.C. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 2013, 41, 4699–4708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, Y.; Johnson, M.A.; Lidder, P.; Vogel, J.T.; van Erp, H.; Green, P.J. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 2004, 328, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- Forieri, I.; Sticht, C.; Reichelt, M.; Gretz, N.; Hawkesford, M.J.; Malagoli, M.; Wirtz, M.; Hell, R. System analysis of metabolism and the transcriptome in Arabidopsis thaliana roots reveals differential co-regulation upon iron, sulfur and potassium deficiency. Plant Cell Environ. 2017, 40, 95–107. [Google Scholar] [CrossRef]
- Forieri, I.; Wirtz, M.; Hell, R. Toward new perspectives on the interaction of iron and sulfur metabolism in plants. Front. Plant Sci. 2013, 4, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Song, Z.; Sun, Z. GO molecular function coding based protein subcellular localization prediction. Chin. Sci. Bull. 2007, 52, 2240–2245. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Matsui, A.; Tanaka, M.; Mizunashi, K.; Nakaminami, K.; Hayashi, M.; Iida, K.; Toyoda, T.; Nguyen, D.V.; Seki, M. Loss of Arabidopsis 5’-3’ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 2015, 56, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Rymarquis, L.A.; Souret, F.F.; Green, P.J. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 2011, 17, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Von Roretz, C.; Di Marco, S.; Mazroui, R.; Gallouzi, I.E. Turnover of AU-rich-containing mRNAs during stress: A matter of survival. Wiley Interdisc. Rev. RNA 2011, 2, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Bannai, H.; Tamada, Y.; Maruyama, O.; Nakai, K.; Miyano, S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 2002, 18, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Hubstenberger, A.; Courel, M.; Benard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 2017, 68, 144–157.e145. [Google Scholar] [CrossRef]
- Takeo, K.; Ito, T. Subcellular localization of VIP1 is regulated by phosphorylation and 14-3-3 proteins. FEBS Lett. 2017, 591, 1972–1981. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-∆∆C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Briesemeister, S.; Blum, T.; Brady, S.; Lam, Y.; Kohlbacher, O.; Shatkay, H. SherLoc2: A high-accuracy hybrid method for predicting subcellular localization of proteins. J. Proteome Res. 2009, 8, 5363–5366. [Google Scholar] [CrossRef] [PubMed]
- Pierleoni, A.; Martelli, P.L.; Fariselli, P.; Casadio, R. BaCelLo: A balanced subcellular localization predictor. Bioinformatics 2006, 22, e408–e416. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Briesemeister, S.; Rahnenführer, J.; Kohlbacher, O. Yloc—An interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010, 38, W497–W502. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armbruster, L.; Uslu, V.V.; Wirtz, M.; Hell, R. The Recovery from Sulfur Starvation Is Independent from the mRNA Degradation Initiation Enzyme PARN in Arabidopsis. Plants 2019, 8, 380. https://doi.org/10.3390/plants8100380
Armbruster L, Uslu VV, Wirtz M, Hell R. The Recovery from Sulfur Starvation Is Independent from the mRNA Degradation Initiation Enzyme PARN in Arabidopsis. Plants. 2019; 8(10):380. https://doi.org/10.3390/plants8100380
Chicago/Turabian StyleArmbruster, Laura, Veli Vural Uslu, Markus Wirtz, and Rüdiger Hell. 2019. "The Recovery from Sulfur Starvation Is Independent from the mRNA Degradation Initiation Enzyme PARN in Arabidopsis" Plants 8, no. 10: 380. https://doi.org/10.3390/plants8100380
APA StyleArmbruster, L., Uslu, V. V., Wirtz, M., & Hell, R. (2019). The Recovery from Sulfur Starvation Is Independent from the mRNA Degradation Initiation Enzyme PARN in Arabidopsis. Plants, 8(10), 380. https://doi.org/10.3390/plants8100380





