Efficient Characterization of Tetraploid Watermelon
Abstract
:1. Introduction
2. Results
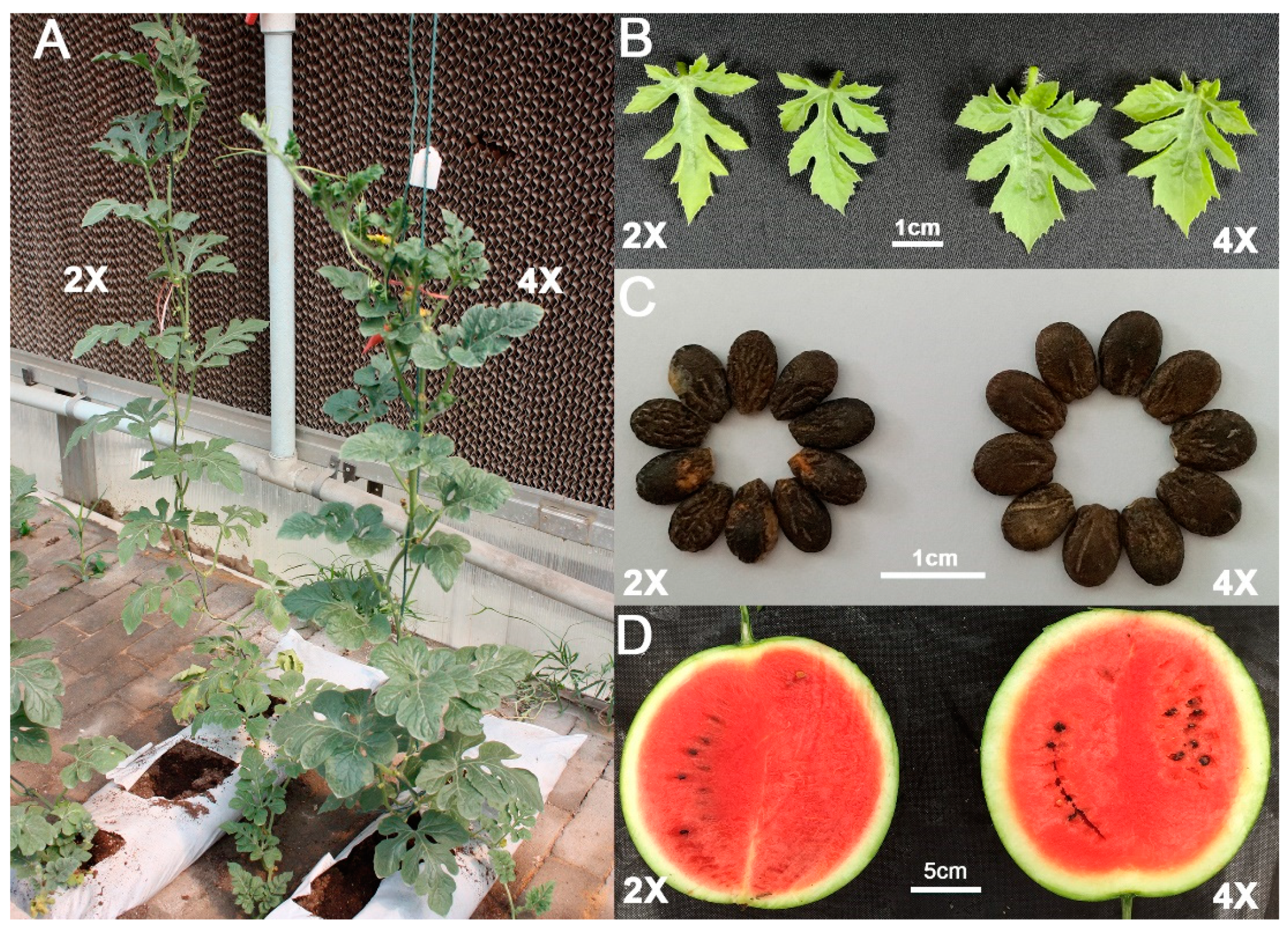
2.1. Tetraploid Induction and Morphological Characterization in Watermelon
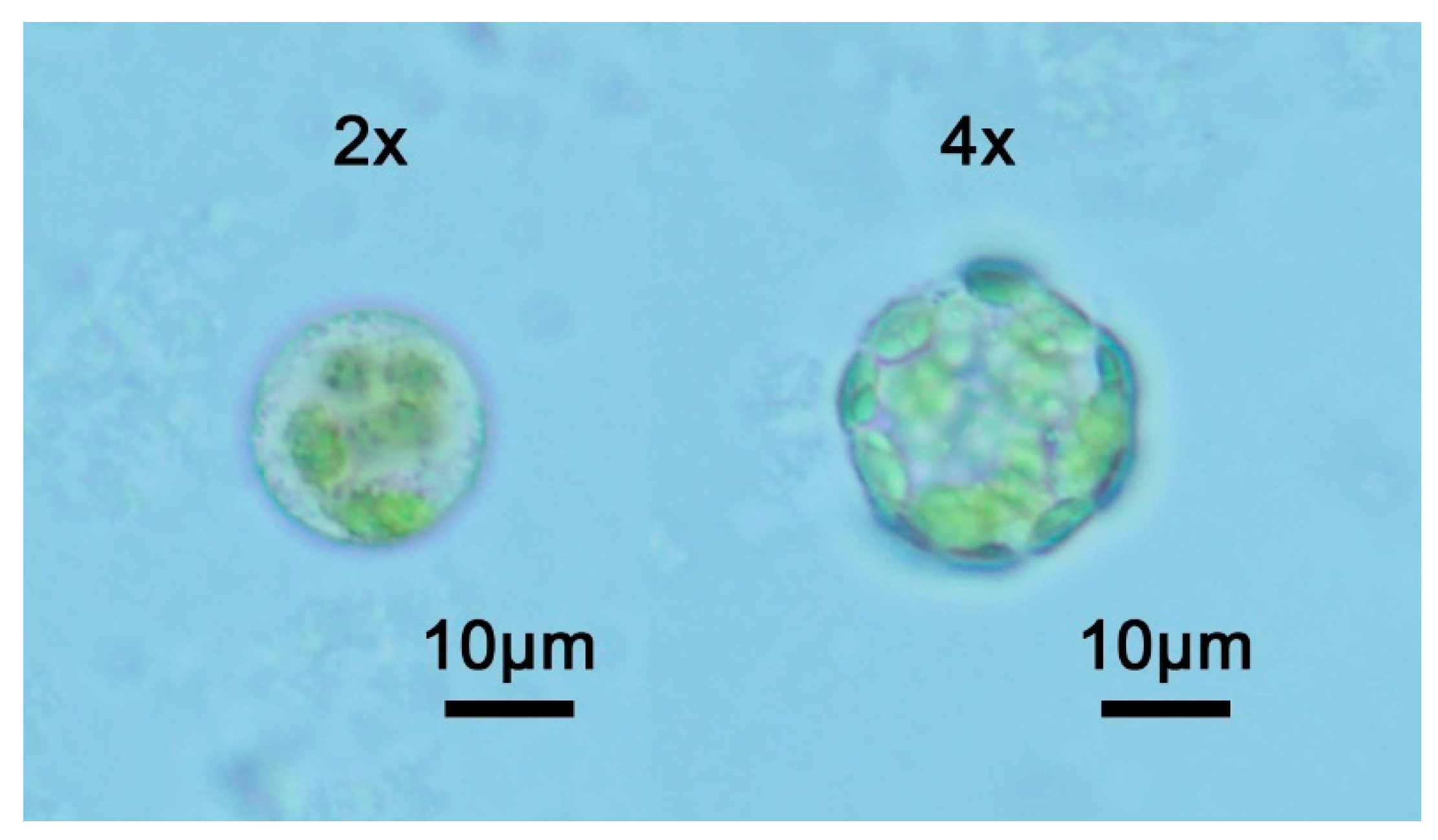
2.2. Tetraploid Characterization by Flow Cytometric Analysis
2.3. 5S rDNA Sequences in Watermelon Genome
2.4. Tetraploid Characterization by RT-qPCR
3. Discussion
3.1. Method for Identification of Watermelon Polyploid
3.2. RT-qPCR Could be Used for Tetraploid Characterization
4. Materials and Methods
4.1. Tetraploid Induction and Morphological Characterization
4.2. Flow Cytometric Analysis
4.3. 5S rDNA Sequence Retrieval
4.4. Protoplast and DNA Isolation
4.5. RT-qPCR Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blakeslee, A.; Avery, A.G. Methods of inducing doubling of chromosomes in plants: By treatment with colchicine. J. Hered. 1937, 28, 393–411. [Google Scholar] [CrossRef]
- Soltis, D.E.; Misra, B.B.; Shan, S.; Chen, S.; Soltis, P.S. Polyploidy and the proteome. Biochim. Biophys. Acta 2016, 1864, 896–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfree, R.C.; Marshall, D.J.; Young, A.G.; Miller, C.H.; Mathews, S. Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. R. Soc. Open Sci. 2017, 4, 170934. [Google Scholar] [CrossRef] [PubMed]
- Jaskani, M.J.; Kwon, S.W.; Kim, D.H. Comparative study on vegetative, reproductive and qualitative traits of seven diploid and tetraploid watermelon lines. Euphytica 2005, 145, 259–268. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, S.; Cheng, Z.; Wan, X.; Yan, Z.; King, S. Lycopene and citrulline contents in watermelon (Citrullus lanatus) fruit with different ploidy and changes during fruit development. Acta Hortic. 2010, 871, 543–550. [Google Scholar]
- Zhu, H.; Zhao, S.; Lu, X.; He, N.; Gao, L.; Dou, J.; Bie, Z.; Liu, W. Genome duplication improves the resistance of watermelon root to salt stress. Plant Physiol. Biochem. 2018, 133, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.; Huang, X.Q.; Liu, J.W.; Liu, W.G. Raising the frequency of inducing tetraploid watermelon by treating with colchicine. Acta Hort 1995, 402, 18–22. [Google Scholar]
- Zhang, X. Tetraploid Watermelons Producing Small Fruits. U.S. Patent 8742208, 6 March 2014. [Google Scholar]
- Compton, M.E.; Gray, D.J. Shoot organogenesis on cotyledons of watermelon. Hortscience A Publ. Am. Soc. Hortic. Sci. 1991, 26, 772. [Google Scholar]
- Krug, M.G.Z.; Stipp, L.C.L.; Rodriguez, A.P.M.; Mendes, B.M.J. In vitro organogenesis in watermelon cotyledons. Pesqui. Agropecuária Bras. 2005, 40, 1678–3921. [Google Scholar] [CrossRef]
- Compton, M.E.; Gray, D.J.; Elmstrom, G.W. Identification of tetraploid regenerants from cotyledons of diploid watermelon culturedin vitro. Euphytica 1996, 87, 165–172. [Google Scholar] [CrossRef]
- Rhodes, B.; Zhang, X.P. Hybrid seed production in watermelon. New Seed 1999, 1, 69–88. [Google Scholar] [CrossRef]
- McCuistion, F.; Elmstrom, G.W. Identifying polyploids of various cucurbits by various guard cell chloroplast numbers. Proc. Fla. State Hort. Soc. 1993, 106, 155–157. [Google Scholar]
- Sari, N.; Abak, K.; Pitrat, M. Comparison of ploidy level screening methods in watermelon: Citrullus lanatus (Thunb.) Matsum. and Nakai. Sci. Hortic. Amst. 1999, 82, 265–277. [Google Scholar] [CrossRef]
- Fahleson, J.; Dixelius, J.; Sundberg, E.; Glimelius, K. Correlation between flow cytometric determination of nuclear DNA content and chromosome number in somatic hybrids within Brassiceae. Plant Cell Rep. 1988, 7, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ming, W.; Yan, A. AFLP Analysis of the Genetic Diversity between Diploid and Autopoly-ploidy Watermelon. J. Fruit Sci. 2004, 21, 46–49. [Google Scholar]
- Fox, M.H.; Galbraith, D.W. Application of flow cytometry and sorting to higher plant systems. Flow Cytom. Sorting 1990, 633–650. [Google Scholar]
- Arumuganathan, K.; Earle, E.D. Estimation of nuclear DNA content of plants by flow cutometry. Plant Mol. Biol. Report. 1991, 9, 221–231. [Google Scholar] [CrossRef]
- Koh, G.C. Tetraploid production of Moodeungsan watermelon. J. Kor. Soc. Hortic. Sci 2002, 43, 671–676. [Google Scholar]
- Jaskani, M.J.; Kwon, S.W.; Koh, G.C.; Huh, Y.C.; Ko, B.R. Induction and characterization of tetraploid watermelon. J. Kor. Soc. Hort. Sci 2004, 45, 60–65. [Google Scholar]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Rubio-Piña, J.; Quiroz-Moreno, A.; Sánchez-Teyer, L.F. A quantitative PCR approach for determining the ribosomal DNA copy number in the genome of Agave tequila Weber. Electron. J. Biotechnol. 2016, 22, 9–15. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.D.; Leitch, I.J. Nuclear DNA amounts in angiosperms: Progress, problems and prospects. Ann. Bot. 2005, 95, 45–90. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wang, X.; Xiao, F.; Zhou, Y.; Wei, L. Polyploidy Induction and Identification of Hibiscus syriacus. J. West China For. Sci. 2019, 48, 119–125. [Google Scholar]
- Norrmann, G.; Quarin, C.; Keeler, K. Evolutionary implications of meiotic chromosome behavior, reproductive biology and hybridization in 6× and 9×cytotypes of Andropogon gerardii (Poaceae). Am. J. Bot. 1997, 84, 201–208. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Luo, F.X.; Liu, L.; Guo, F. In vitro induction of tetraploids in crape myrtle (Lagerstroemia indica L.). Plant Cell Tissue Organ Cult. 2010, 101, 41–47. [Google Scholar] [CrossRef]
- Leus, L.; Van Laere, K.; Dewitte, A.; Van Huylenbroeck, J. Flow clytometry for plant breeding. Acta Hortic. 2009, 836, 221–226. [Google Scholar] [CrossRef]
- Väinölä, A. Polyploidization and early screening of Rhododendron hybrids. Euphytica 2000, 112, 239–244. [Google Scholar] [CrossRef]
- Montijn, M.B.; Houtsmuller, A.B.; Ten Hoopen, R.; Oud, J.L.; Nanninga, N. The 5S rRNA gene clusters have a defined orientation toward the nucleolus in Petunia hybrida and Crepis capillaries. Chromosome Res. 1999, 7, 387–399. [Google Scholar] [CrossRef]
- De Laat, A.M.M.; Göhde, W.; Vogelzang, M.J.D.C. Determination of ploidy of single plants and plants population by flow cytometry. Plant Breed. 1987, 99, 303–307. [Google Scholar] [CrossRef]
- Brown, S.C.; Devaux, P.; Marie, D.; Bergounioux, C.; Petit, P.X. Cytométrie enflux: Application à l’analyse de la ploidie chez les végétaux. Biofuture 1991, 105, 2–16. [Google Scholar]
- Dhar, M.K.; Kaul, S.; Friebe, B.; Gill, B.S. Chromosome identification in Plantago ovata Forsk. Through C-banding and FISH. Curr. Sci. 2002, 83, 150–152. [Google Scholar]
- Murata, M.; Heslop-Harrison, J.S.; Motoyoshi, F. Physical mapping of the 5S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clones. Plant J. 1997, 12, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zeng, H.X.; Shi, X.F.; Ren, J.; Cheng, W.S.; Yang, Y.X.; Li, Y.H.; Sun, Y.H. Selection of Tetraploid of a Yellow Flesh Mini-Watermelon Using Oryzalin. Adv. Mater. Res. 2013, 838, 2449–2454. [Google Scholar] [CrossRef]
- Huang, X.; Chen, J.; Bao, Y.; Liu, L.; Jiang, H.; An, X.; Dai, L.; Wang, B.; Peng, D. Transcript Profiling Reveals Auxin and Cytokinin Signaling Pathways and Transcription Regulation during In Vitro Organogenesis of Ramie (Boehmeria nivea L. Gaud). PLoS ONE 2014, 9, e113768. [Google Scholar] [CrossRef]




| Genotype | Fruit | Seed Area (cm2) | Protoplast Area (μm2) | ||
|---|---|---|---|---|---|
| Vertical Diameter (cm) | Cross Diameter (cm) | Aspect Ratio | |||
| E46 | 16.44 ± 0.61 | 16.08 ± 0.44 | 1.02 ± 0.02 | 0.30 ± 0.01b | 258.06 ± 10.87b |
| yE46 | 17.66 ± 0.23 | 17.84 ± 0.68 | 0.99 ± 0.03 | 0.37 ± 0.01a | 591.02 ± 83.11a |
| ID | Chromosome | Location | Length (bp) |
|---|---|---|---|
| 5s1 | Chr0 | 8960268–8960378(+) | 110 |
| 5s2 | Chr0 | 8960616–8960726(+) | 110 |
| 5s3 | Chr1 | 20672252–20672358(+) | 106 |
| 5s4 | Chr1 | 20672524–20672634(+) | 110 |
| 5s5 | Chr1 | 20672797–20672907(+) | 110 |
| 5s6 | Chr1 | 20673032–20673142(+) | 110 |
| 5s7 | Chr1 | 20673317–20673424(+) | 107 |
| 5s8 | Chr1 | 20673590–20673700(+) | 110 |
| 5s9 | Chr1 | 20674311–20674421(+) | 110 |
| 5s10 | Chr1 | 20674587–20674697(+) | 110 |
| 5s11 | Chr1 | 20675133–20675243(+) | 110 |
| 5s12 | Chr1 | 20675409–20675519(+) | 110 |
| 5s13 | Chr1 | 20676875–20676985(+) | 110 |
| 5s14 | Chr1 | 20677151–20677261(+) | 110 |
| 5s15 | Chr1 | 20677427–20677537(+) | 110 |
| 5s16 | Chr1 | 20677612–20677722(+) | 110 |
| 5s17 | Chr1 | 20677885–20677995(+) | 110 |
| 5s18 | Chr1 | 20678161–20678271(+) | 110 |
| 5s19 | Chr4 | 11951587–11951479(−) | 108 |
| 5s20 | Chr7 | 30808653–30808545(−) | 108 |
| 5s21 | Chr8 | 16056311–16056201(−) | 110 |
| 5s22 | Chr9 | 23004927–23005033(+) | 102 |
| 5s23 | Chr11 | 1932259–1932150(−) | 110 |
| Sample | NanoDrop | qPCR | ||||
|---|---|---|---|---|---|---|
| DNA Conc. | Ploidy | Ct | Total Amount of DNA | DNA Conc. | Ploidy | |
| E46 | 110.7 ng/μL | 2 × | 24.0873 | 21.4740 ng | 1.0737 ng/μL | 2 × |
| yE46 | 196.1 ng/μL | 4 × | 23.6052 | 36.4961 ng | 1.8248 ng/μL | 4 × |
| Gene | Forward 5′–3′ | Reverse 5′–3′ | Size (bp) | Temperature (°C) | Efficiency % |
|---|---|---|---|---|---|
| 5S | CGATCATACCAGCACTAAAGCACC | ATGCAACACGAGGACTTCCCAG | 111 | 60 | 104 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Bao, Y.; Xie, Z.; Huang, X.; Sun, Y.; Feng, G.; Zeng, H.; Ren, J.; Li, Y.; Xiong, J.; et al. Efficient Characterization of Tetraploid Watermelon. Plants 2019, 8, 419. https://doi.org/10.3390/plants8100419
Zhang N, Bao Y, Xie Z, Huang X, Sun Y, Feng G, Zeng H, Ren J, Li Y, Xiong J, et al. Efficient Characterization of Tetraploid Watermelon. Plants. 2019; 8(10):419. https://doi.org/10.3390/plants8100419
Chicago/Turabian StyleZhang, Na, Yaning Bao, Zhouli Xie, Xing Huang, Yuhong Sun, Gang Feng, Hongxia Zeng, Jian Ren, Yuhua Li, Jianshun Xiong, and et al. 2019. "Efficient Characterization of Tetraploid Watermelon" Plants 8, no. 10: 419. https://doi.org/10.3390/plants8100419
APA StyleZhang, N., Bao, Y., Xie, Z., Huang, X., Sun, Y., Feng, G., Zeng, H., Ren, J., Li, Y., Xiong, J., Chen, W., Yan, C., & Tang, M. (2019). Efficient Characterization of Tetraploid Watermelon. Plants, 8(10), 419. https://doi.org/10.3390/plants8100419





