Isolation and Characterization of Avirulent and Virulent Strains of Agrobacterium tumefaciens from Rose Crown Gall in Selected Regions of South Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Samples
2.2. Gall Extraction
2.3. Isolation and Phenotypic Characterization of Agrobacterium Isolates
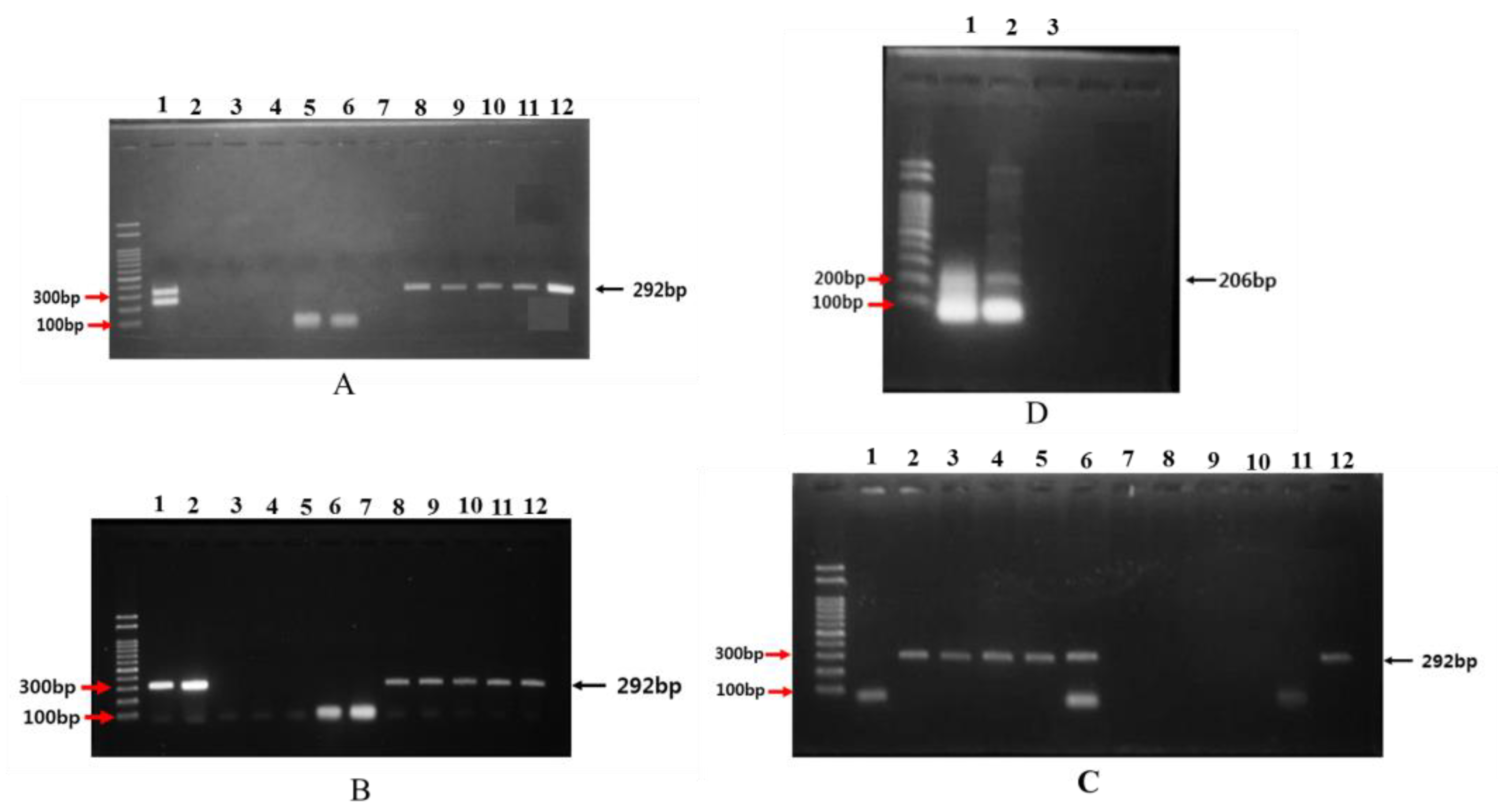
2.4. Analysis of 16S rDNA for Bacteria Isolates
2.4.1. PCR Amplification of 16S rDNA
2.4.2. Sequence Analysis of 16S rDNA
2.5. Enzyme Assay (API ZYM strip) for Utilization Test of Carbon Sources
2.6. Motility Assay
2.7. Bioassay for Testing Pathogenicity
2.8. Biofilm Assay
2.9. PCR for Genes Related to Carbon Source Utilization
3. Results and Discussion
3.1. Isolation and Phenotypic Characterization
3.2. Biofilm Formation
3.3. Tumor Formation
3.4. Genes Associated with Opine Metabolism
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sahin, F.; Aysan, Y. An Outbreak of Crown Gall Disease on Rose Caused by Agrobacterium Tumefaciens in Turkey. Plant Pathol. 2003, 52, 780. [Google Scholar]
- Tzfira, T.; Citovsky, V. Agrobacterium-Mediated Genetic Transformation of Plants: Biology and Biotechnology. Curr. Opin. Biotechnol. 2006, 17, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Gordon, J.E. The Agrobacterium Ti Plasmids. Microbiol. Spectr. 2014, 2, 1–18. [Google Scholar]
- Pacurar, D.I.; Thordal-Christensen, H.; Pacurar, M.L.; Pamfil, D.; Botez, C.; Bellini, C. Agrobacterium Tumefaciens: From Crown Gall Tumors to Genetic Transformation. Physiol. Mol. Plant Pathol. 2011, 76, 76–81. [Google Scholar] [CrossRef]
- Guyon, P.; Chilton, M.D.; Petit, A.; Tempe, J. Agropine in "Null-Type" Crown Gall Tumors: Evidence for Generality of the Opine Concept. Proc. Natl. Acad. Sci. USA 1980, 77, 2693–2697. [Google Scholar] [CrossRef]
- Palanichelvam, K.; Veluthambi, K. Octopine-And Nopaline-Inducible Proteins in Agrobacterium Tumefaciens are Also Induced by Arginine. Curr. Microbiol. 1996, 33, 156–162. [Google Scholar] [CrossRef]
- Dessaux, Y.; Petit, A.; Tempe, J. Chemistry and Biochemistry of Opines, Chemical Mediators of Parasitism. Phytochemistry 1993, 34, 31–38. [Google Scholar] [CrossRef]
- Petit, A.; David, C.; Dahl, G.A.; Ellis, J.G.; Guyon, P.; Casse-Delbart, F.; Tempe, J. Further Extension of the Opine Concept: Plasmids in Agrobacterium Rhizogenes Cooperate for Opine Degradation. Mol. Gen. Genet. 1983, 190, 204–214. [Google Scholar] [CrossRef]
- Spaink, H.P.; Kondorosi, A.; Hooykaas, P. The Rhizobiaceae—Molecular Biology of Model Plant Associated Bacteria; Kluwer: Dordrecht, The Netherland; Springer Science and Business Media: Berlin, Germany, 1998. [Google Scholar]
- Kim, H.S.; Yi, H.; Myung, J.; Piper, K.R.; Farrand, S.K. Opine-Based Agrobacterium Competitiveness: Dual Expression Control of the Agrocinopine Catabolism (ACC) Operon by Agrocinopines and Phosphate Levels. J. Bacteriol. 2008, 190, 3700–3711. [Google Scholar] [CrossRef]
- Palanichelvam, K.; Oger, P.; Clough, S.J.; Cha, C.; Bent, A.F.; Farrand, S.K. A Second T-Region of the Soybean-Supervirulent Chrysopine-Type Ti Plasmid pTiChry5, and Construction of a Fully Disarmed Vir Helper Plasmid. Mol. Plant Microbe Interact. 2000, 13, 1081–1091. [Google Scholar] [CrossRef]
- Britton, M.T.; Escobar, M.A.; Dandekar, A.M. The Oncogenes of Agrobacterium Tumefaciens and Agrobacterium Rhizogenes. In Agrobacterium: From Biology to Biotechnology; Springer Science and Business Media LLC: Berlin, Germany, 2008; pp. 523–563. [Google Scholar]
- Gohlke, J.; Deeken, R. Plant Responses to Agrobacterium Tumefaciens and Crown Gall Development. Front. Plant Sci. 2014, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Dessaux, Y.; Faure, D. Quorum Sensing and Quorum Quenching in Agrobacterium: A Go/No Go System. Genes 2018, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanovic, N.; Pulawska, J. Evolutionary Relatedness and Classification of Tumor-Inducing and Opine-Catabolic Plasmids in Three Rhizobium Rhizogenes Strains Isolated from the Same Crown Gall Tumor. Genome Boil. Evol. 2019, 11, 1525–1540. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Oger, P.M.; Schrammeijer, B.; Hooykaas, P.J.J.; Farrand, S.K.; Winans, S.C. The Bases of Crown Gall Tumorigenesis. J. Bacteriol. 2000, 182, 3885–3895. [Google Scholar] [CrossRef] [Green Version]
- Habeeb, L.F. Transcription of the Octopine Catabolism Operon of the Agrobacterium Tumor-Inducing Plasmid pTiA6 Is Activated by a LysR-Type Regulatory Protein. Mol. Plant Microbe Interact. 1991, 4, 379. [Google Scholar] [CrossRef]
- Wang, L.; Winans, S.C. High Angle and Ligand-Induced Low Angle DNA Bends Incited by OccR Lie in the Same Plane with OccR Bound to the Interior Angle. J. Mol. Boil. 1995, 253, 32–38. [Google Scholar] [CrossRef]
- Von Bodman, S.B.; Hayman, G.T.; Farrand, S.K. Opine Catabolism and Conjugal Transfer of the Nopaline Ti Plasmid pTiC58 are Coordinately Regulated by a Single Repressor. Proc. Natl. Acad. Sci. USA 1992, 89, 643–647. [Google Scholar] [CrossRef]
- White, C.E.; Winans, S.C. Cell–Cell Communication in the Plant Pathogen Agrobacterium Tumefaciens. Philos. Trans. R. Soc. B Boil. Sci. 2007, 362, 1135–1148. [Google Scholar] [CrossRef]
- Venturi, V.; Fuqua, C. Chemical Signaling Between Plants and Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 2013, 51, 17–37. [Google Scholar] [CrossRef]
- Luo, Z.Q.; Qin, Y.; Farrand, S.K. The Antiactivator TraM Interferes with the Autoinducer-Dependent Binding of TraR to DNA by Interacting with the C-Terminal Region of the Quorum-Sensing Activator. J. Boil. Chem. 2000, 275, 7713–7722. [Google Scholar] [CrossRef]
- Haudecoeur, E.; Faure, D. A Fine Control of Quorum-Sensing Communication in Agrobacterium Tumefaciens. Commun. Integr. Boil. 2010, 3, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Zhu, J.; Winans, S.C. TrlR, a Defective TraR-Like Protein of Agrobacterium Tumefaciens, Blocks TraR Function in Vitro by Forming Inactive TrlR:TraR Dimers. Mol. Microbiol. 2001, 40, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Laverde-Gomez, J.A.; Sarkar, M.K.; Christie, P.J. Regulation of Bacterial Type IV Secretion Systems. In Regulation of Bacterial Virulence; Vasil, M., Darwin, A., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 335–362. [Google Scholar]
- Morton, E.R.; Fuqua, C. Phenotypic Analyses of Agrobacterium. Curr. Protoc. Microbiol. 2012, 3, 3D.3. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Hiraishi, A. Direct Automated Sequencing of 16S rDNA Amplified by Polymerase Chain Reaction from Bacterial Cultures Without DNA Purification. Lett. Appl. Microbiol. 1992, 15, 210–213. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput rRNA Analysis. Nucl. Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Plewniak, F.; Poch, O. A Comprehensive Comparison of Multiple Sequence Alignment Programs. Nucleic Acids Res. 1999, 27, 2682–2690. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Boil. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Boil. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Adler, J.; Templeton, B. The Effect of Environmental Conditions on the Motility of Escherichia coli. J. Gen. Microbiol. 1967, 46, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloni, R.; Wolf, A.; Feigenbaum, P.; Avni, A.; Klee, H.J. The Never Ripe Mutant Provides Evidence That Tumor-Induced Ethylene Controls the Morphogenesis of Agrobacterium Tumefaciens-Induced Crown Galls on Tomato Stems. Plant Physiol. 1998, 117, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.S.; Yabuki, J.; Matsumoto, S.; Kageyama, K.; Fukui, H. PCR Primers for Identification of Opine Types of Agrobacterium Tumefaciens in Japan. J. Gen. Plant Pathol. 2003, 69, 258–266. [Google Scholar] [CrossRef]
- Pulawska, J. Crown Gall of Stone Fruits and Nuts, Economic Significance and Diversity of its Causal Agents: Tumorigenic Agrobacterium Spp. J. Plant Pathol. 2010, 92, S87–S98. [Google Scholar]
- Tiwary, B.N.; Prasad, B.; Ghosh, A.; Kumar, S.; Jain, R.K. Characterization of Two Novel Biovar of Agrobacterium Tumefaciens Isolated from Root Nodules of Vicia faba. Curr. Microbiol. 2007, 55, 328–333. [Google Scholar] [CrossRef]
- Pionnat, S.; Keller, H.; Hericher, D.; Bettachini, A.; Dessaux, Y.; Nesme, X.; Poncet, C. Ti Plasmids from Agrobacterium Characterize Rootstock Clones That Initiated a Spread of Crown Gall Disease in Mediterranean Countries. Appl. Environ. Microbiol. 1999, 65, 4197–4206. [Google Scholar]
- Kalogeraki, V.S.; Winans, S.C. The Octopine-Type Ti Plasmid pTiA6 of Agrobacterium Tumefaciens Contains a Gene Homologous to the Chromosomal Virulence gene acvB. J. Bacteriol. 1995, 177, 892–897. [Google Scholar] [CrossRef]
- Pansegrau, W.; Schoumacher, F.; Hohn, B.; Lanka, E. Site-Specific Cleavage and Joining of Single-Stranded DNA by VirD2 Protein of Agrobacterium Tumefaciens Ti Plasmids: Analogy to Bacterial Conjugation. Proc. Natl. Acad. Sci. USA 1993, 90, 11538–11542. [Google Scholar] [CrossRef]
- Haas, J.H.; Moore, L.W.; Ream, W.; Manulis, S. Universal PCR Primers for Detection of Phytopathogenic Agrobacterium Strains. Appl. Environ. Microbiol. 1995, 61, 2879–2884. [Google Scholar]
- Subramoni, S.; Nathoo, N.; Klimov, E.; Yuan, Z.C. Agrobacterium Tumefaciens Responses to Plant-Derived Signaling Molecules. Front. Plant Sci. 2014, 5, 322. [Google Scholar] [CrossRef]
- Kim, H.; Farrand, S.K. Opine Catabolic Loci from Agrobacterium Plasmids Confer Chemotaxis to Their Cognate Substrates. Mol. Plant Microbe Interact. 1998, 11, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Oger, P.; Farrand, S.K. Two Opines Control Conjugal Transfer of an Agrobacterium Plasmid by Regulating Expression of Separate Copies of the Quorum-Sensing Activator Gene traR. J. Bacteriol. 2002, 184, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Piper, K.R.; Von Bodman, S.B.; Hwang, I.; Farrand, S.K. Hierarchical Gene Regulatory Systems Arising From Fortuitous Gene Associations: Controlling Quorum Sensing by the Opine Regulon in Agrobacterium. Mol. Microbiol. 1999, 32, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Oger, P.; Farrand, S.K. Co-Evolution of the Agrocinopine Opines and the Agrocinopine-Mediated Control of TraR, the Quorum-Sensing Activator of the Ti Plasmid Conjugation System. Mol. Microbiol. 2001, 41, 1173–1185. [Google Scholar] [CrossRef]
- Melnyk, C.W. Connecting the Plant Vasculature to Friend or Foe. New Phytol. 2017, 213, 1611–1617. [Google Scholar] [CrossRef]
- Garfinkel, D.J.; Nester, E.W. Agrobacterium Tumefaciens Mutants Affected in Crown Gall Tumorigenesis and Octopine Catabolism. J. Bacteriol. 1980, 144, 732–743. [Google Scholar]
- Vicedo, B.; Penalver, R.; Asins, M.J.; Lopez, M.M. Biological Control of Agrobacterium Tumefaciens, Colonization, and pAgK84 Transfer with Agrobacterium Radiobacter K84 and the Tra Mutant Strain K1026. Appl. Environ. Microbiol. 1993, 59, 309–315. [Google Scholar]
- Lang, J.; Vigouroux, A.; Planamente, S.; El Sahili, A.; Blin, P.; Aumont-Nicaise, M.; Dessaux, Y.; Morera, S.; Faure, D. Agrobacterium Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host. PLOS Pathog. 2014, 10, e1004444. [Google Scholar] [CrossRef]
- Kado, C.I. Historical Account on Gaining Insights on the Mechanism of Crown Gall Tumorigenesis Induced by Agrobacterium Tumefaciens. Front. Microbiol. 2014, 5, 340. [Google Scholar] [CrossRef]
- Klapwijk, P.M.; Scheulderman, T.; Schilperoort, R.A. Coordinated Regulation of Octopine Degradation and Conjugative Transfer of Ti Plasmids in Agrobacterium Tumefaciens: Evidence for a Common Regulatory Gene and Separate Operons. J. Bacteriol. 1978, 136, 775–785. [Google Scholar]
- Tzfira, T.; Citovsky, V. (Eds.) Agrobacterium: From Biology to Biotechnolog; Springer-Verlag: New York, NY, USA, 2008. [Google Scholar]
- Khan, S.R.; Su, S.; Farrand, S.K. Degradation of Acyl-HSLs by AttM Lactonase and its Role in Controlling the Conjugative Transfer of Ti-Plasmids in Agrobacterium Tumefaciens. Plasmid 2007, 4, 217. [Google Scholar]
- Dessaux, Y.; Guyon, P.; Petit, A.; Tempe, J.; Demarez, M.; Legrain, C.; Tate, M.E.; Farrand, S.K. Opine Utilization by Agrobacterium Spp.: Octopine-Type Ti Plasmids Encode Two Pathways for Mannopinic Acid Degradation. J. Bacteriol. 1988, 170, 2939–2946. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Kado, C.I. Ti Plasmid and Chromosomal Ornithine Catabolism Genes of Agrobacterium Tumefaciens C58. J. Bacteriol. 1983, 155, 196–202. [Google Scholar] [PubMed]
- Nester, E. Beyond My Wildest Expectations. Annu. Rev. Microbiol. 2014, 68, 1–20. [Google Scholar] [CrossRef]
- Kim, K.S.; Baek, C.H.; Lee, J.K.; Yang, J.M.; Farrand, S.K. Intracellular Accumulation of Mannopine, an Opine Produced by Crown Gall Tumors, Transiently Inhibits Growth of Agrobacterium Tumefaciens. Mol. Plant Microbe Interact. 2001, 14, 793–803. [Google Scholar] [CrossRef] [PubMed]



| Gene | Primer Name | Primer Sequence | Reference |
|---|---|---|---|
| Agrocinopine | ACC-F | 5’ AGGAATGAAAATGAACCCTCT 3’ | In this study |
| ACC-R | 5’ CTCCGAACTGAACCAACTCCC 3’ | ||
| Nopaline | RB-F | 5’ TGACAGGATATATTGGCGGGTAA 3’ | [36] |
| RB-R | 5’ TGCTCCTCCGTCAGGCTTTCCGA 3’ | ||
| Octopine | OCS-F | 5’ ATGGCTAAAGTGGCAATTTTGGG 3’ | [36] |
| OCS-R | 5’ TCAGATTGAASTTCGCCAACTCG 3’ |
| Enzyme | Agrobacterium tumefaciens Strains a | ||||
|---|---|---|---|---|---|
| RC b 053 | RC b 084 | RC b 111 | RC c 081 | RC c 178 | |
| Alkaline phosphatase | − | + | + | + | + |
| Esterase | + | + | + | + | + |
| Esterase lipase | + | + | + | + | + |
| Lipase | + | − | + | − | + |
| Leucine arylamidase | − | − | + | + | + |
| Valine arylamidase | + | + | − | + | − |
| Cystine arylamidase | − | + | + | − | + |
| Trypsin | − | − | + | + | + |
| a-chymotrypsin | + | − | + | − | − |
| Acid phosphatase | + | + | + | + | + |
| Naphthol-AS-BI-phosphohydrolase | + | + | + | + | + |
| a-galactosidase | − | − | + | − | + |
| b-galactosidase | + | + | + | + | − |
| b-glucuronidase | − | − | + | − | + |
| a-glucosidase | − | − | + | − | + |
| b-glucosidase | − | + | + | + | + |
| N-acetyl-b-glucosaminidase | + | − | + | − | + |
| a-mannosidase | − | − | + | − | + |
| a-fucosidase | + | − | − | − | − |
| Strain a | Abs b. (Mean ± std.) | Gall formed c (+, −) | Strain | Abs. (Mean ± std.) | Path (+, −) | Strain | Abs. (Mean ± std.) | Gall formed (+, −) |
|---|---|---|---|---|---|---|---|---|
| C58 | 0.49 ± 0.13 | + | RC076 | 0.11 ± 0.01 | − | RC132 | 0.21 ± 0.04 | + |
| RC002 | 0.78 ± 0.15 | − | RC077 | 0.10 ± 0.01 | − | RC133 | 0.12 ± 0.01 | + |
| RC003 | 0.61 ± 0.23 | − | RC079 | 0.13 ± 0.04 | − | RC134 | 0.18 ± 0.05 | + |
| RC004 | 0.61 ± 0.18 | − | RC080 | 0.12 ± 0.01 | − | RC135 | 0.27 ± 0.01 | + |
| RC005 | 0.61 ± 0.04 | − | RC081 | 0.23 ± 0.00 | − | RC140 | 0.18 ± 0.03 | + |
| RC006 | 0.61 ± 0.13 | − | RC082 | 0.32 ± 0.16 | − | RC141 | 0.85 ± 0.15 | − |
| RC007 | 0.71 ± 0.11 | − | RC083 | 0.09 ± 0.01 | − | RC160 | 0.61 ± 0.11 | − |
| RC008 | 0.42 ± 0.03 | + | RC084 | 0.27 ± 0.04 | − | RC162 | 0.14 ± 0.04 | − |
| RC009 | 0.26 ± 0.04 | + | RC085 | 0.14 ± 0.01 | − | RC165 | 0.12 ± 0.02 | − |
| RC011 | 0.14 ± 0.02 | + | RC087 | 0.07 ± 0.01 | − | RC170 | 0.09 ± 0.01 | + |
| RC012 | 0.61 ± 0.18 | + | RC088 | 0.14 ± 0.02 | − | RC171 | 0.11 ± 0.02 | − |
| RC013 | 0.55 ± 0.11 | + | RC089 | 0.11 ± 0.02 | − | RC172 | 0.08 ± 0.01 | − |
| RC014 | 0.56 ± 0.07 | + | RC090 | 0.13 ± 0.01 | − | RC173 | 0.10 ± 0.01 | − |
| RC016 | 0.60 ± 0.08 | − | RC091 | 0.16 ± 0.02 | − | RC174 | 0.09 ± 0.01 | − |
| RC017 | 0.10 ± 0.01 | − | RC092 | 0.11 ± 0.03 | − | RC175 | 0.10 ± 0.02 | − |
| RC018 | 0.53 ± 0.30 | − | RC093 | 0.12 ± 0.02 | − | RC178 | 0.48 ± 0.05 | + |
| RC019 | 0.17 ± 0.01 | − | RC096 | 0.25 ± 0.04 | − | RC179 | 0.11 ± 0.01 | − |
| RC026 | 0.09 ± 0.00 | + | RC096 | 0.25 ± 0.04 | − | RC180 | 0.15 ± 0.02 | − |
| RC027 | 0.22 ± 0.03 | + | RC098 | 0.18 ± 0.03 | − | |||
| RC029 | 0.11 ± 0.01 | + | RC099 | 0.08 ± 0.00 | − | |||
| RC030 | 0.08 ± 0.01 | + | RC101 | 0.10 ± 0.01 | − | |||
| RC032 | 0.17 ± 0.02 | + | RC103 | 0.10 ± 0.00 | − | |||
| RC033 | 0.13 ± 0.03 | − | RC111 | 0.20 ± 0.05 | − | |||
| RC036 | 0.16 ± 0.02 | − | RC112 | 0.87 ± 0.61 | − | |||
| RC049 | 0.13 ± 0.00 | − | RC113 | 0.12 ± 0.00 | − | |||
| RC053 | 0.11 ± 0.01 | − | RC114 | 0.20 ± 0.06 | − | |||
| RC066 | 0.97 ± 0.21 | − | RC116 | 0.35 ± 0.09 | − | |||
| RC067 | 0.84 ± 0.31 | − | RC117 | 0.16 ± 0.01 | − | |||
| RC068 | 0.81 ± 0.15 | − | RC122 | 0.20 ± 0.01 | − | |||
| RC069 | 0.15 ± 0.01 | − | RC122 | 0.20 ± 0.01 | − | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandrasekaran, M.; Lee, J.M.; Ye, B.-M.; Jung, S.M.; Kim, J.; Kim, J.-W.; Chun, S.C. Isolation and Characterization of Avirulent and Virulent Strains of Agrobacterium tumefaciens from Rose Crown Gall in Selected Regions of South Korea. Plants 2019, 8, 452. https://doi.org/10.3390/plants8110452
Chandrasekaran M, Lee JM, Ye B-M, Jung SM, Kim J, Kim J-W, Chun SC. Isolation and Characterization of Avirulent and Virulent Strains of Agrobacterium tumefaciens from Rose Crown Gall in Selected Regions of South Korea. Plants. 2019; 8(11):452. https://doi.org/10.3390/plants8110452
Chicago/Turabian StyleChandrasekaran, Murugesan, Jong Moon Lee, Bee-Moon Ye, So Mang Jung, Jinwoo Kim, Jin-Won Kim, and Se Chul Chun. 2019. "Isolation and Characterization of Avirulent and Virulent Strains of Agrobacterium tumefaciens from Rose Crown Gall in Selected Regions of South Korea" Plants 8, no. 11: 452. https://doi.org/10.3390/plants8110452








