Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Specimen Collection
2.2. Morphology-Based Species Identification
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Nucleotide Sequence Data Analysis
2.5. Genetic Distance and Phylogenetic Analysis
3. Results
3.1. Taxonomic Resolution of DNA Barcoding Markers
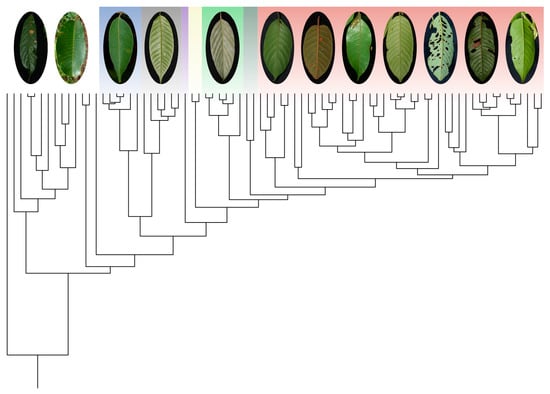
3.2. Species Assignment Using Phylogenetic Trees
4. Discussion
4.1. Applicability of DNA Barcoding
4.2. Phylogenetic Relationships of Sumatran Dipterocarpaceae
4.3. Combined Identification Using Morphology and DNA Barcodes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Indrioko, S.; Gailing, O.; Finkeldey, R. Molecular phylogeny of Dipterocarpaceae in Indonesia based on chloroplast DNA. Plant Syst. Evol. 2006, 261, 99–115. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef] [Green Version]
- Heckenhauer, J.; Samuel, R.; Ashton, P.S.; Abu Salim, K.; Paun, O. Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreeae (Dipterocarpaceae). Mol. Phylogenetics Evol. 2018, 127, 1–13. [Google Scholar] [CrossRef]
- Ashton, P.S. Dipterocarpaceae. In Flowering Plants—Dicotyledons: Malvales, Capparales and Non-Betalain Caryophyllales; Springer: Berlin/Heidelberg, Germany, 2003; Volume 5, pp. 182–197. [Google Scholar]
- Londono, A.C.; Alvarez, E.; Forero, E.; Morton, C.M. A new genus and species of Dipterocarpaceae from the Neotropics. Introduction, Taxonomy, Ecology, and Distribution. Brittonia 1995, 47, 225. [Google Scholar] [CrossRef]
- Brearley, F.Q.; Banin, L.F.; Saner, P. The ecology of the Asian dipterocarps. Plant Ecol. Divers 2016, 9, 429–436. [Google Scholar] [CrossRef]
- Slik, J.W.F.; Poulsen, A.D.; Ashton, P.S.; Cannon, C.H.; Eichhorn, K.A.O.; Kartawinata, K.; Lanniari, I.; Nagamasu, H.; Nakagawa, M.; van Nieuwstadt, M.G.L.; et al. A floristic analysis of the lowland dipterocarp forests of Borneo. J. Biogeogr. 2003, 30, 1517–1531. [Google Scholar] [CrossRef]
- Surianegara, I.C.; Lemmens, R.H.M.J. Timber trees: Major commercial timbers. In Plant Resources of South-East Asia; van der Maesen, L.J.G., Somaatmadja, S., Eds.; ESCAP CGPRT Centre: Bogor, Indonesia, 1990. [Google Scholar]
- Ang, C.C.; O’Brien, M.J.; Ng, K.K.S.; Lee, P.C.; Hector, A.; Schmid, B.; Shimizu, K.K. Genetic diversity of two tropical tree species of the Dipterocarpaceae following logging and restoration in Borneo: high genetic diversity in plots with high species diversity. Plant Ecol. Divers 2016, 9, 459–469. [Google Scholar] [CrossRef]
- Avise, J. Evolutionary Pathways in Nature a Phylogenetic Approach; Cambridge University Press: New York, NY, USA, 2006. [Google Scholar]
- Foxworthy, F.W. Dipterocarpaceae of the Malay Peninsula; Malayan Forest Records; Federated Malay States Government, Forest Department: Kepong, Malaysia, 1932; p. 10.
- Symington, C.F.; Ashton, P.S.; Appanah, S. Foresters’ manual of dipterocarps; Forest Research Institute Malaysia (FRIM): Kuala Lumpur, Malaysia, 2004. [Google Scholar]
- Ashton, P.S. Dipterocarpaceae. In Flora Malesiana I; Martinus Nijhoff Publishers: Hague, The Netherlands, 1982; Volume 9, pp. 237–552. [Google Scholar]
- Gamage, D.T.; de Silva, M.P.; Inomata, N.; Yamazaki, T.; Szmidt, A.E. Comprehensive molecular phylogeny of the subfamily Dipterocarpoideae (Dipterocarpaceae) based on chloroplast DNA sequences. Genes Genet. Syst. 2006, 81, 1–12. [Google Scholar] [CrossRef]
- Harnelly, E.; Thomy, Z.; Fathiya, N. Phylogenetic analysis of Dipterocarpaceae in Ketambe Research Station, Gunung Leuser National Park (Sumatra, Indonesia) based on rbcL and matK genes. Biodiversitas J. Biol. Divers 2018, 19, 1074–1080. [Google Scholar] [CrossRef]
- Yulita, K.S.; Bayer, R.J.; West, J.G. Molecular phylogenetic study of Hopea and Shorea (Dipterocarpaceae): Evidence from the trnL–trnF and internal transcribed spacer regions. Plant Species Biol. 2005, 20, 167–182. [Google Scholar] [CrossRef]
- Kamiya, K.; Harada, K.; Tachida, H.; Ashton, P.S. Phylogeny of PgiC gene in Shorea and its closely related genera (Dipterocarpaceae), the dominant trees in Southeast Asian tropical rain forests. Am. J. Bot. 2005, 92, 775–788. [Google Scholar] [CrossRef]
- Cvetković, T.; Hinsinger, D.D.; Strijk, J.S. Exploring evolution and diversity of Chinese Dipterocarpaceae using next-generation sequencing. Sci. Rep. 2019, 9, 11639. [Google Scholar] [CrossRef]
- Heckenhauer, J.; Paun, O.; Chase, M.W.; Ashton, P.S.; Kamariah, A.S.; Samuel, R. Molecular phylogenomics of the tribe Shoreeae (Dipterocarpaceae) using whole plastid genomes. Ann. Bot. 2019, 123, 857–865. [Google Scholar] [CrossRef]
- Dayanandan, S.; Ashton, P.S.; Williams, S.M.; Primack, R.B. Phylogeny of the tropical tree family Dipterocarpaceae based on nucleotide sequences of the chloroplast rbcL gene. Am. J. Bot. 1999, 86, 1182–1190. [Google Scholar] [CrossRef]
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Maury, G. Diptérocarpacées: du Fruit à la Plantule. Ph.D. Thesis, Université Paul Sabatier, Tolouse, France, 1978. [Google Scholar]
- Heckenhauer, J.; Samuel, R.; Ashton, P.S.; Turner, B.; Barfuss, M.H.J.; Jang, T.-S.; Temsch, E.M.; Mccann, J.; Salim, K.A.; Attanayake, A.M.A.S.; et al. Phylogenetic analyses of plastid DNA suggest a different interpretation of morphological evolution than those used as the basis for previous classifications of Dipterocarpaceae (Malvales). Bot. J. Linn. Soc. 2017, 185, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Heckenhauer, J.; Salim, K.A.; Chase, M.W.; Dexter, K.G.; Pennington, R.T.; Tan, S.; Kaye, M.E.; Samuel, R. Plant DNA barcodes and assessment of phylogenetic community structure of a tropical mixed dipterocarp forest in Brunei Darussalam (Borneo). PLoS ONE 2017, 12, e0185861. [Google Scholar] [CrossRef]
- Amandita, F.Y.; Rembold, K.; Vornam, B.; Rahayu, S.; Siregar, I.Z.; Kreft, H.; Finkeldey, R. DNA barcoding of flowering plants in Sumatra, Indonesia. Ecol. Evol. 2019, 9, 1858–1868. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, M.L.; Andra Clark, A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants: DNA barcoding. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef]
- Webb, C.O.; Slik, J.W.F.; Triono, T. Biodiversity inventory and informatics in Southeast Asia. Biodivers. Conserv. 2010, 19, 955–972. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. PNAS 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Laumonier, Y.; Uryu, Y.; Stüwe, M.; Budiman, A.; Setiabudi, B.; Hadian, O. Eco-floristic sectors and deforestation threats in Sumatra: identifying new conservation area network priorities for ecosystem-based land use planning. Biodivers. Conserv. 2010, 19, 1153–1174. [Google Scholar] [CrossRef]
- IUCN. IUCN Red List Categories and Criteria: Version 3.1, 2nd ed.; IUCN: Gland, Switzerland; Cambridge, UK, 2012.
- Drescher, J.; Rembold, K.; Allen, K.; Beckschäfer, P.; Buchori, D.; Clough, Y.; Faust, H.; Fauzi, A.M.; Gunawan, D.; Hertel, D.; et al. Ecological and socio-economic functions across tropical land-use systems after rainforest conversion. Philos. Trans. R. Soc. B 2016, 371, 20150275. [Google Scholar] [CrossRef]
- Rembold, K.; Mangopo, H.; Tjitrosoedirdjo, S.S.; Kreft, H. Plant diversity, forest dependency, and alien plant invasions in tropical agricultural landscapes. Biol. Conserv. 2017, 213, 234–242. [Google Scholar] [CrossRef]
- Paoletti, A.; Darras, K.; Jayanto, H.; Grass, I.; Kusrini, M.; Tscharntke, T. Amphibian and reptile communities of upland and riparian sites across Indonesian oil palm, rubber and forest. Glob. Ecol. Conserv. 2018, 16, e00492. [Google Scholar] [CrossRef]
- Symington, C.F. Foresters’ Manual of Dipterocarps; Malayan Forest Records; Penerbit Universiti Malaya: Kuala Lumpur, Malaysia, 1943; Volume 16. [Google Scholar]
- Schmitz-Linneweber, C.; Maier, R.M.; Alcaraz, J.-P.; Cottet, A.; Herrmann, R.G.; Mache, R. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001, 45, 307–315. [Google Scholar] [CrossRef]
- Levin, R.A.; Wagner, W.L.; Hoch, P.C.; Nepokroeff, M.; Pires, J.C.; Zimmer, E.A.; Sytsma, K.J. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am. J. Bot. 2003, 90, 107–115. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E.; Smiley, C.J. An rbcL sequence from a Miocene Taxodium (bald cypress). PNAS 1992, 89, 449–451. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.A.R.; Suchard, M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008, 36, 355–368. [Google Scholar] [CrossRef]
- Rambaut, A.; Grass, N.C. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Bioinformatics 1997, 13, 235–238. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer, version 1.5; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2014. [Google Scholar]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: from gene to genome: Plant identification using DNA barcodes. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef]
- Tsumura, Y.; Kado, T.; Yoshida, K.; Abe, H.; Ohtani, M.; Taguchi, Y.; Fukue, Y.; Tani, N.; Ueno, S.; Yoshimura, K.; et al. Molecular database for classifying Shorea species (Dipterocarpaceae) and techniques for checking the legitimacy of timber and wood products. J. Plant. Res. 2011, 124, 35–48. [Google Scholar] [CrossRef]




| Barcode Region | Name of Primer | Primer Sequence (5′ → 3′) | Reference |
|---|---|---|---|
| matK | 3F_KIM_f | CGTACAGTACTTTTGTGTTTACGAG | [28] |
| 1R_KIM_r | ACCCAGTCCATCTGGAAATCTTGGTTC | [28] | |
| 390_f | CGATCTATTCATTCAATATTTC | [28,37] | |
| 990_r | GGACAATGATCCAATCAAGGC | [14] | |
| rbcL | rbcLa_f | ATGTCACCACAAACAGAGACTAAAGC | [38,39] |
| rbcLajf634R_r | GAAACGGTCTCTCCAACGCAT | [40] | |
| trnL-F | B49317_f | CGAAATCGGTAGACGCTACG | [30] |
| B49873_f | GGTTCAAGTCCCTCTATCCC | [30] | |
| A50272_r | ATI’TGAACTGGTGACACGAG | [30] |
| Parameter | matK | rbcL | trnL-F |
|---|---|---|---|
| Number of samples used for amplification and sequencing | 78 | 78 | 78 |
| Number of obtained sequences | 66 | 66 | 42 |
| Sequencing success rate, % | 81.0 | 83.5 | 54.0 |
| Variable sites (proportion), % | 23.1 | 21.3 | 18.0 |
| Parsimony-informative sites (proportion), % | 11.1 | 7.8 | - |
| CG content mean (range), % | 32.3 | 43.7 | 29.0 |
| Length of the alignment, bp | 614 | 603 | 987 |
| Barcode Regions | Species | Genera | Subfamily | Family |
|---|---|---|---|---|
| matK | 17 | 12 | 2 | 1 |
| rbcL | 12 | 11 | 2 | 1 |
| trnL-F | 18 | 14 | 2 | 1 |
| matK + rbcL | 20 | 16 | 2 | 1 |
| matK + rbcL + trnL-F | 21 | 16 | 2 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carneiro de Melo Moura, C.; Brambach, F.; Jair Hernandez Bado, K.; Krutovsky, K.V.; Kreft, H.; Tjitrosoedirdjo, S.S.; Siregar, I.Z.; Gailing, O. Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra. Plants 2019, 8, 461. https://doi.org/10.3390/plants8110461
Carneiro de Melo Moura C, Brambach F, Jair Hernandez Bado K, Krutovsky KV, Kreft H, Tjitrosoedirdjo SS, Siregar IZ, Gailing O. Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra. Plants. 2019; 8(11):461. https://doi.org/10.3390/plants8110461
Chicago/Turabian StyleCarneiro de Melo Moura, Carina, Fabian Brambach, Kevin Jair Hernandez Bado, Konstantin V. Krutovsky, Holger Kreft, Sri Sudarmiyati Tjitrosoedirdjo, Iskandar Z. Siregar, and Oliver Gailing. 2019. "Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra" Plants 8, no. 11: 461. https://doi.org/10.3390/plants8110461
APA StyleCarneiro de Melo Moura, C., Brambach, F., Jair Hernandez Bado, K., Krutovsky, K. V., Kreft, H., Tjitrosoedirdjo, S. S., Siregar, I. Z., & Gailing, O. (2019). Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra. Plants, 8(11), 461. https://doi.org/10.3390/plants8110461