Plant Taxonomic Diversity Better Explains Soil Fungal and Bacterial Diversity than Functional Diversity in Restored Forest Ecosystems
Abstract
:1. Introduction
2. Results
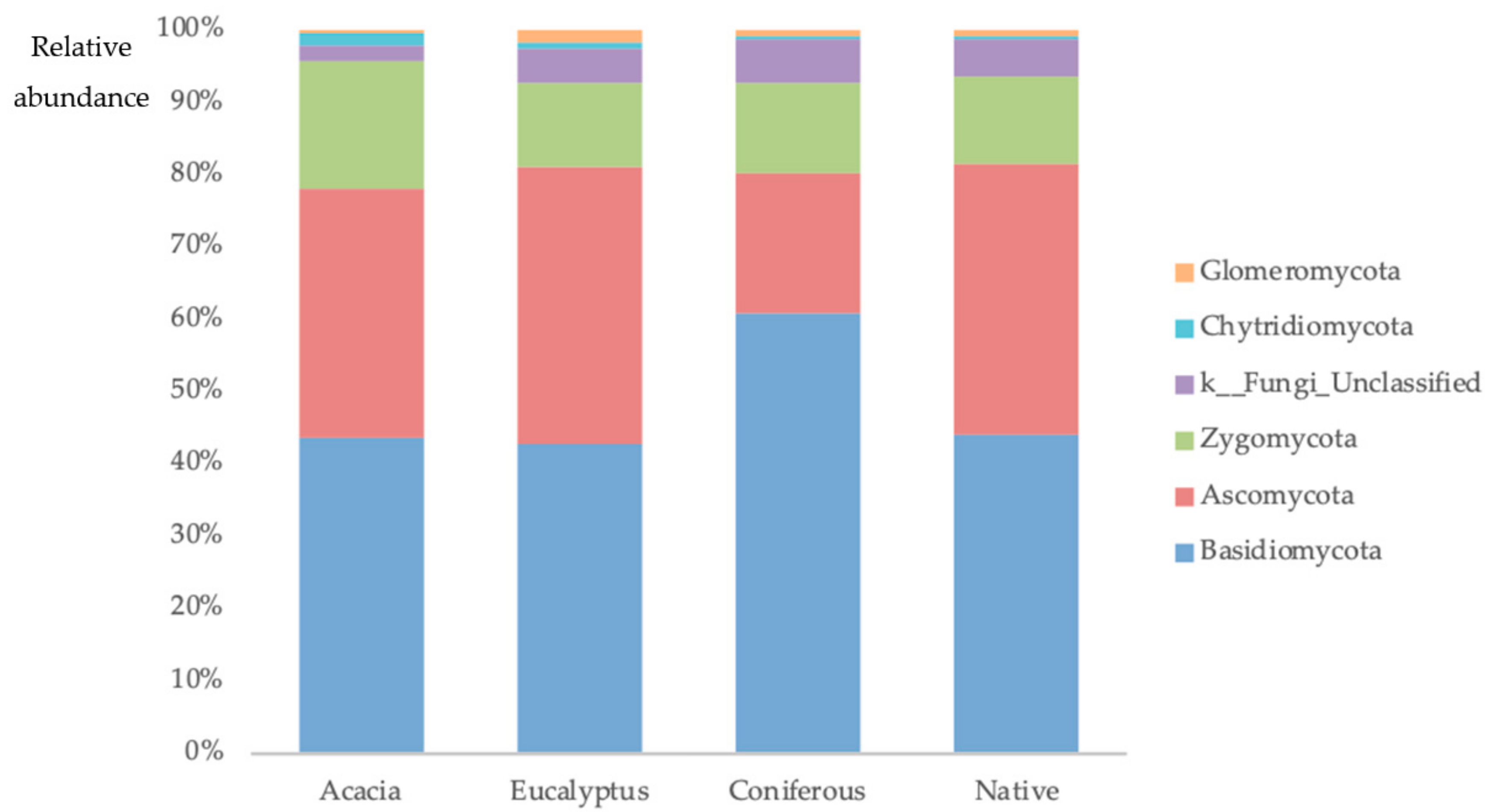
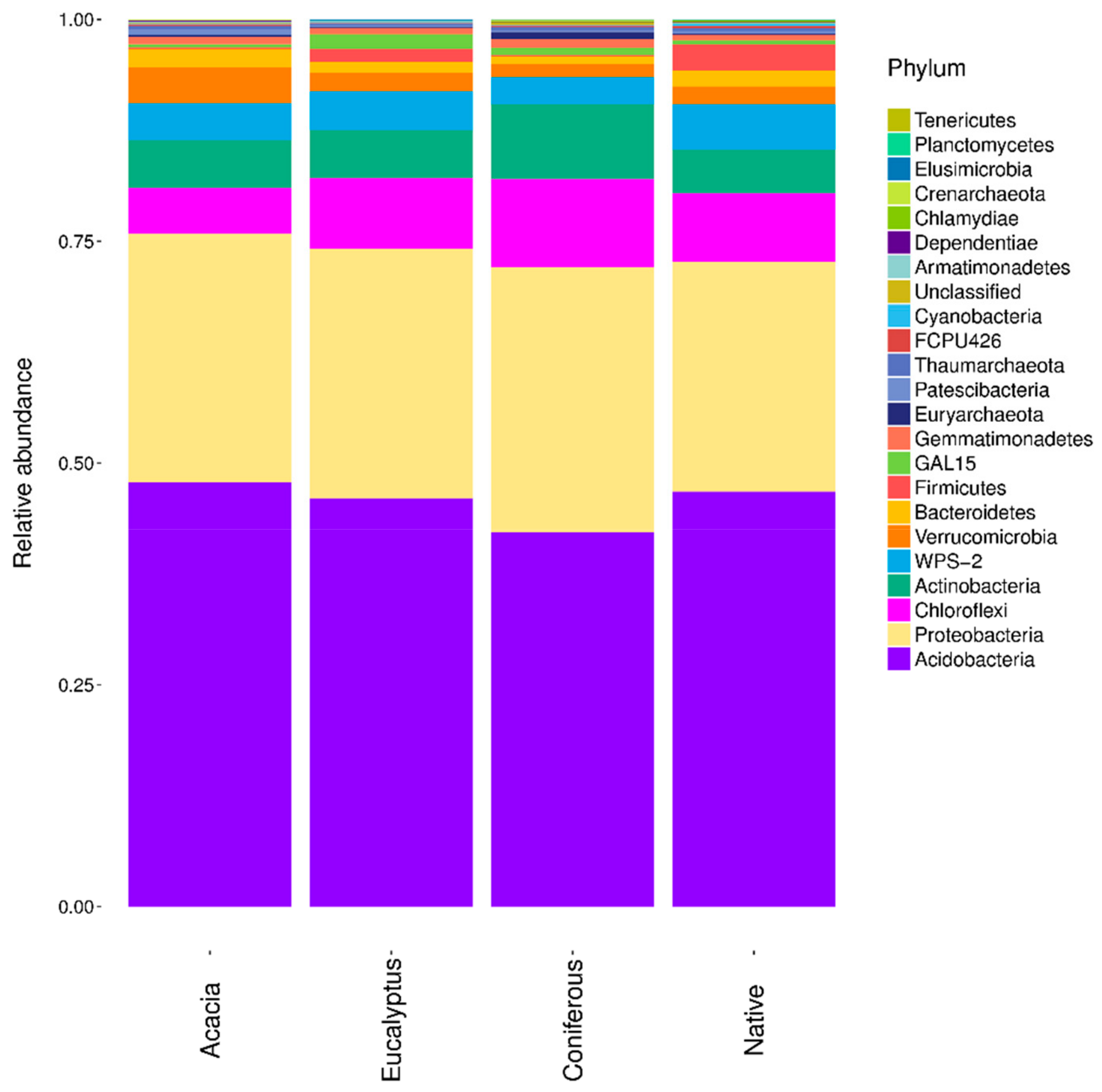
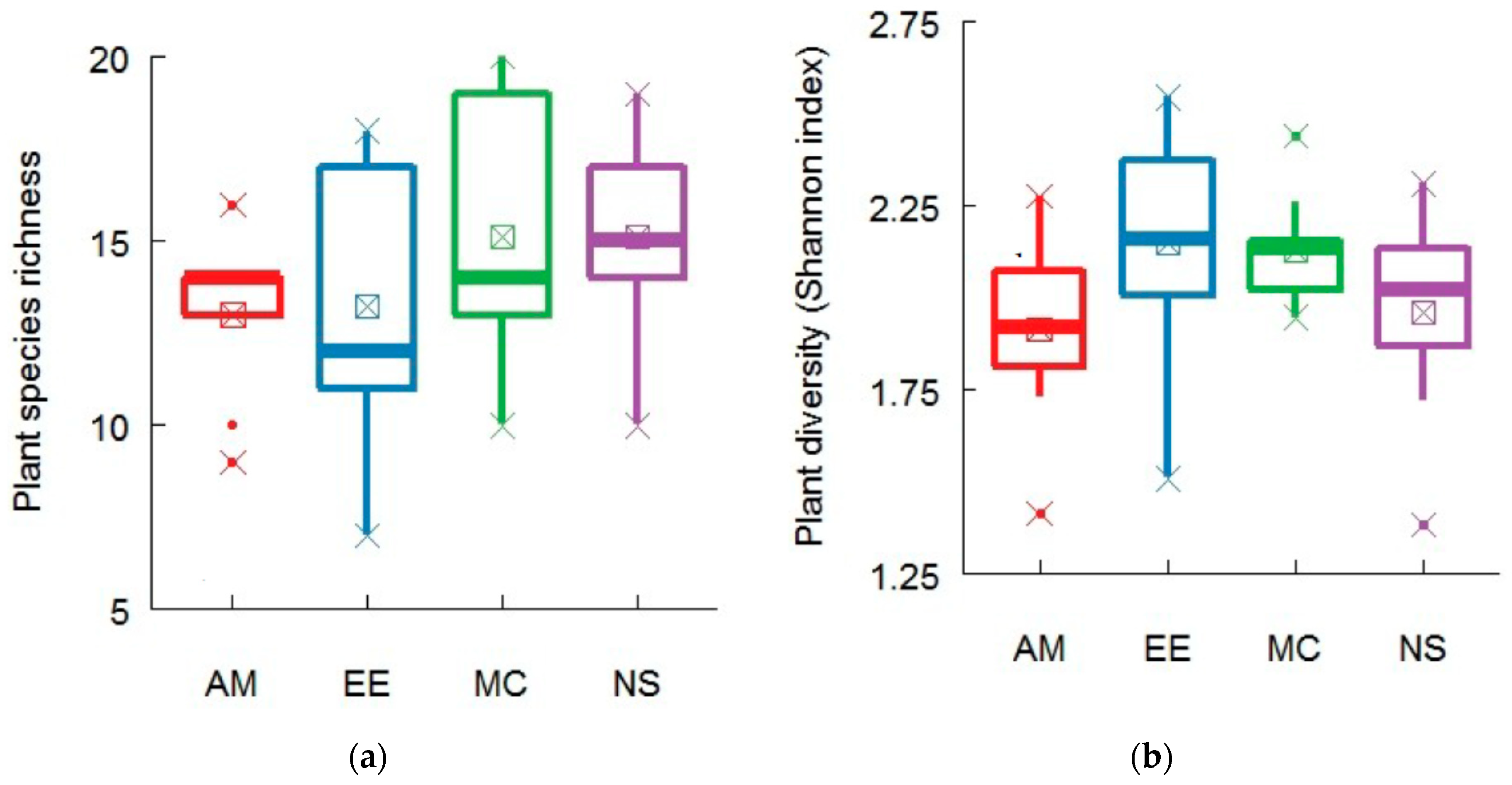
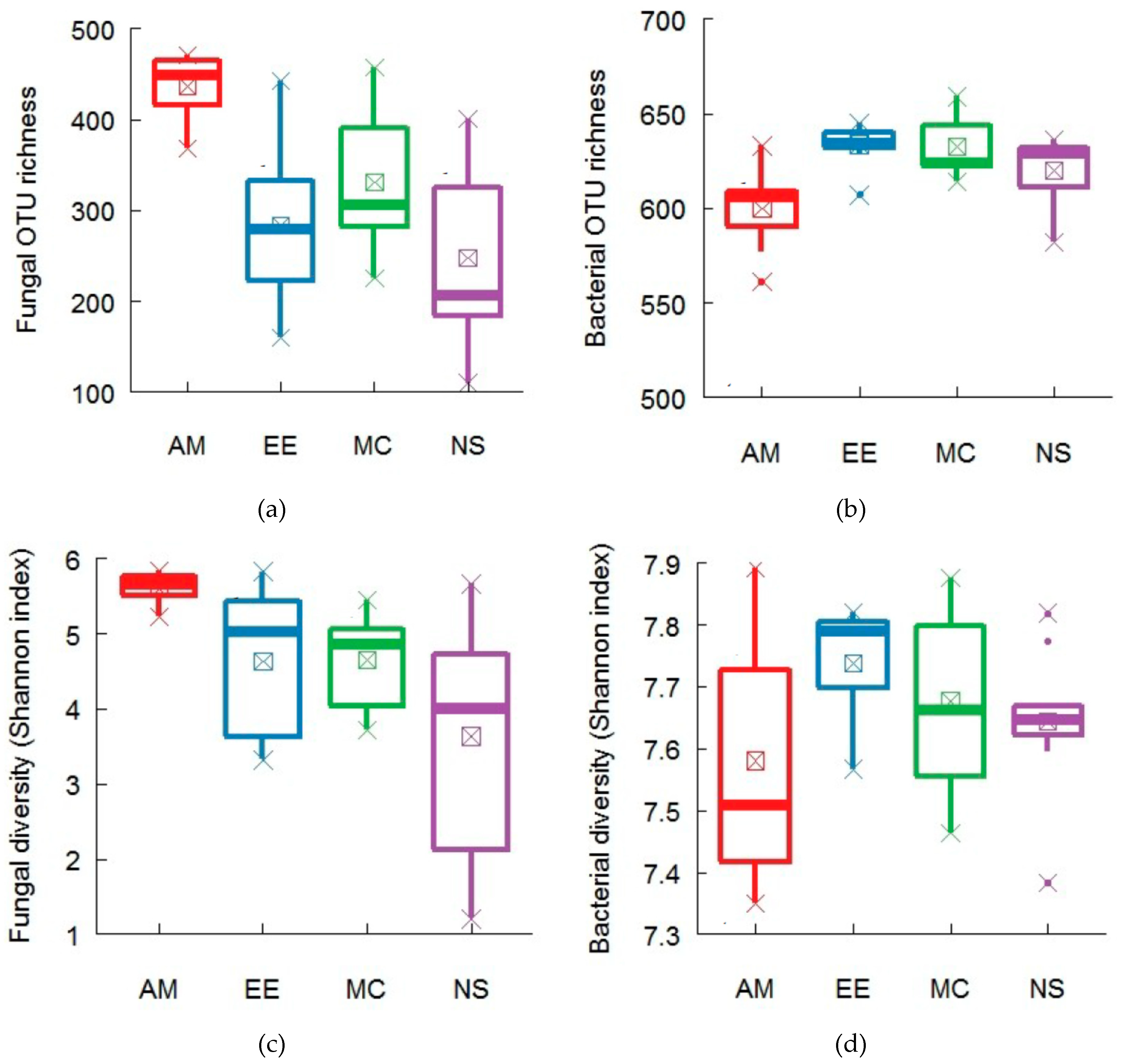
2.1. Plant and Soil Community Composition
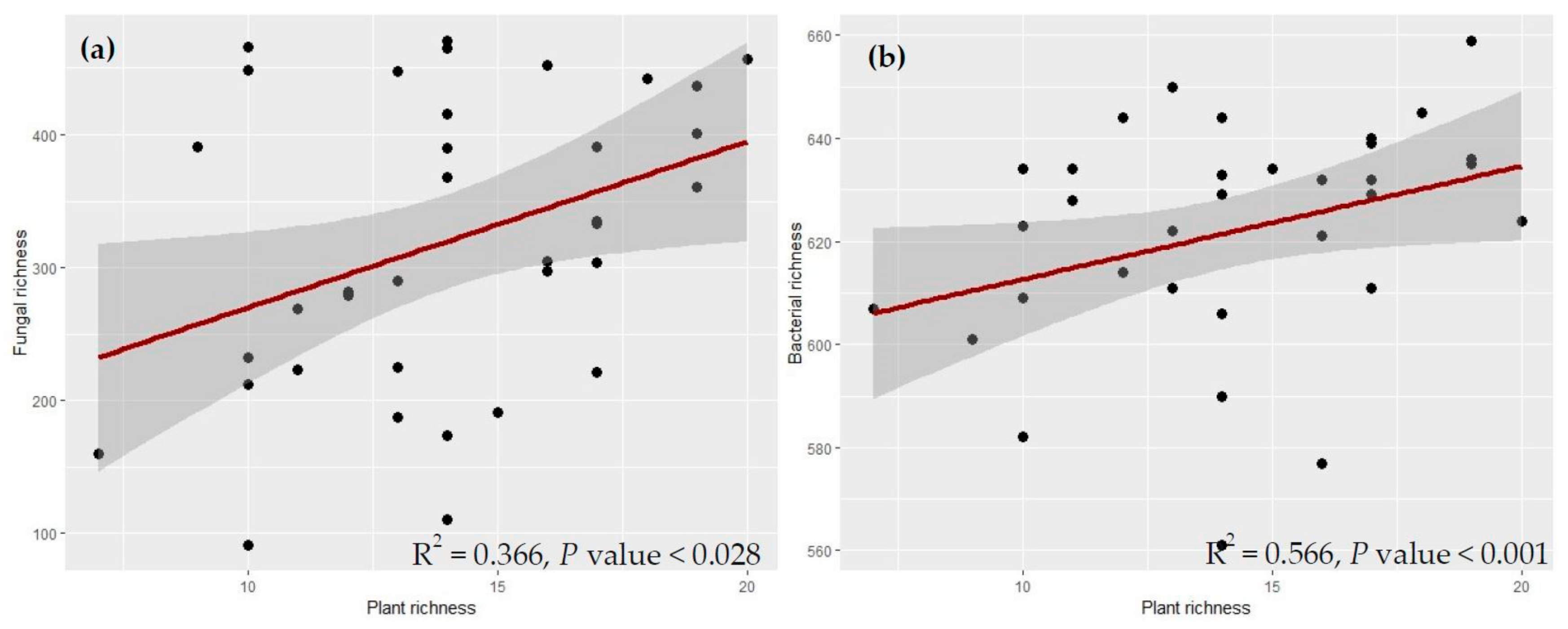
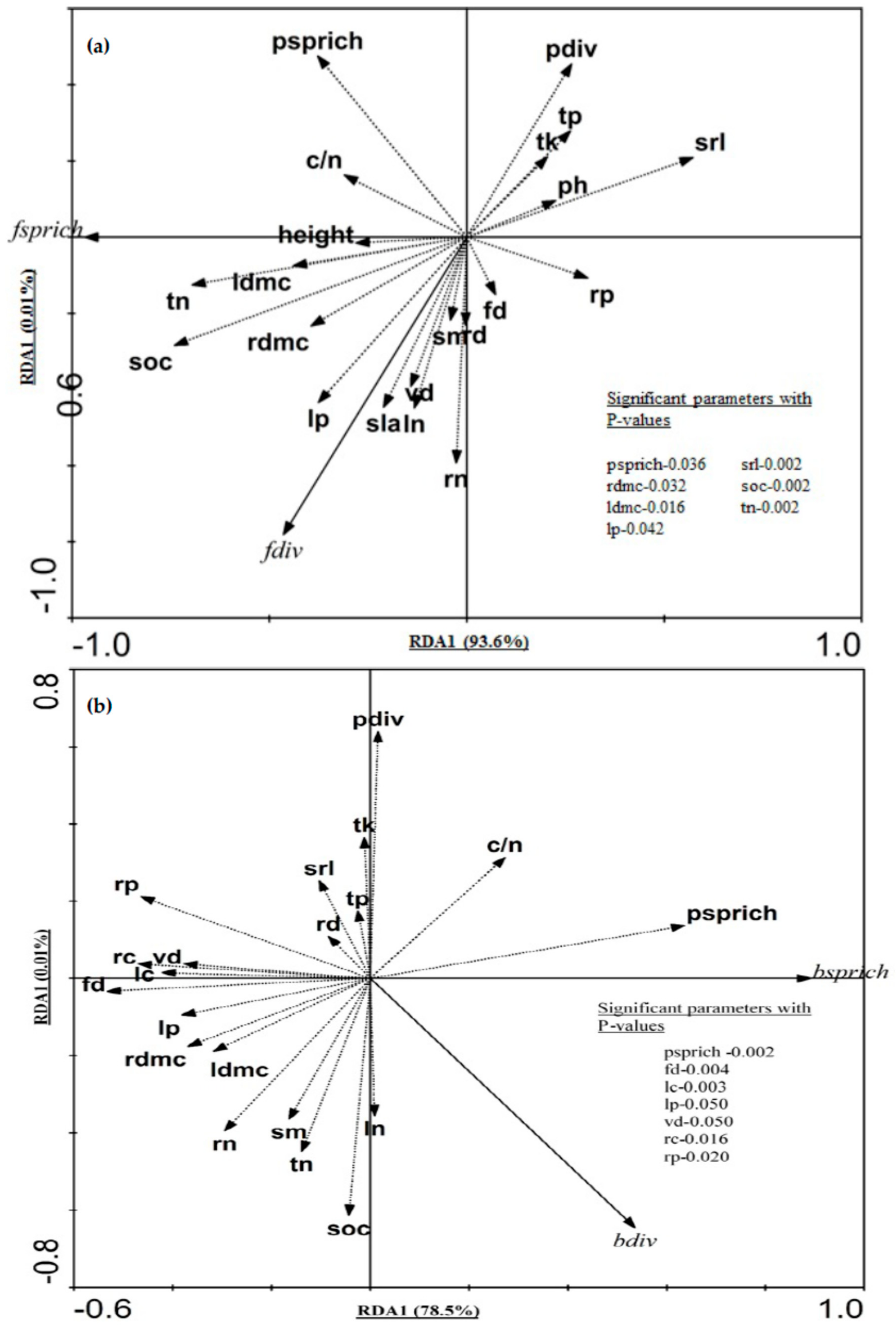
2.2. Relative Contribution of Plant Taxonomic Diversity Versus Functional Diversity in Explaining Fungal and Bacterial Diversity
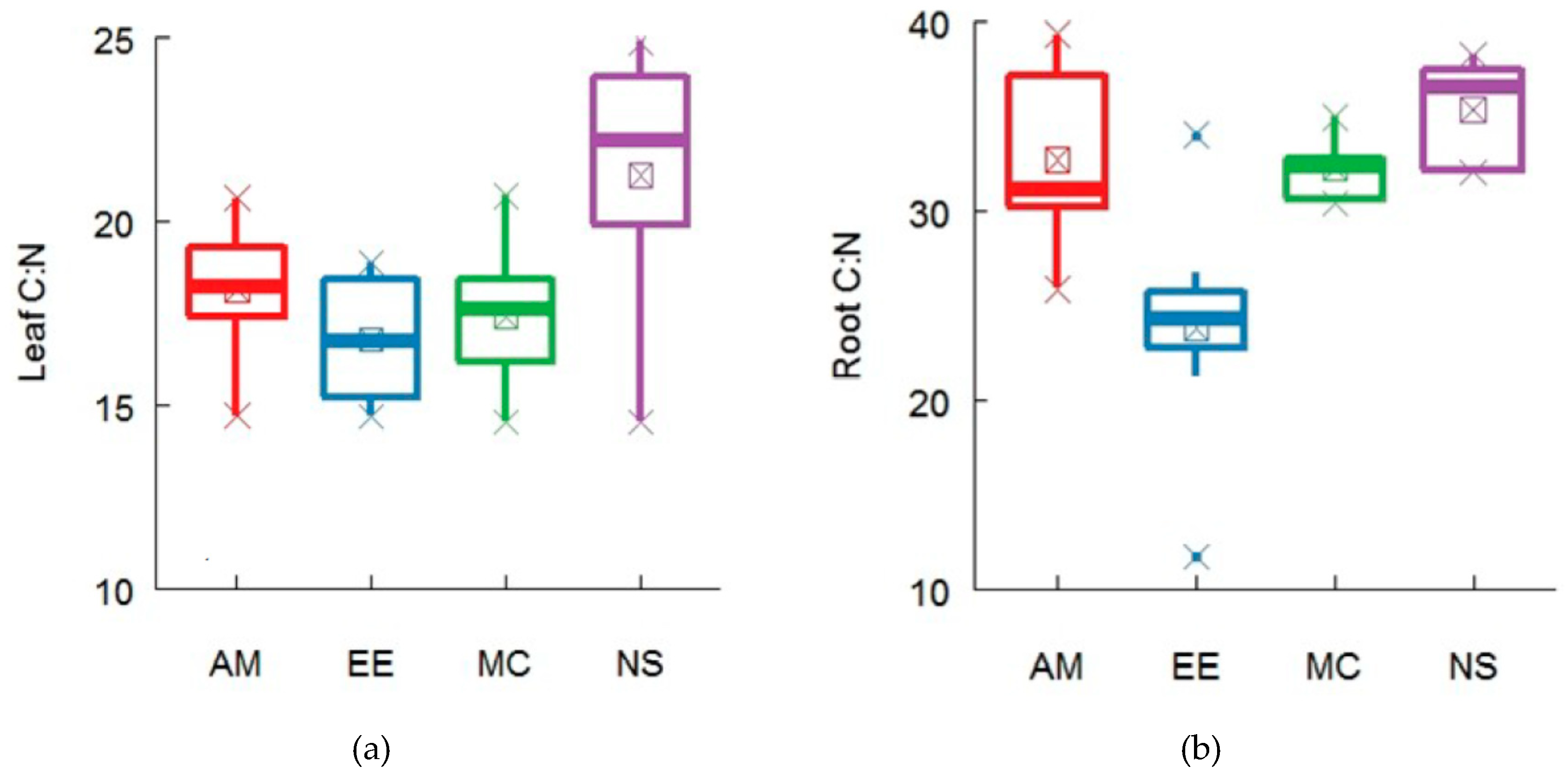
2.3. Functional Traits Variation across Forest Ecosystems and Contribution of Functional Traits for Explaining Microbial Diversity
3. Discussion
4. Materials and Methods
4.1. Experimental Site
4.2. Field Plots and Plant Sampling
4.3. Plant Functional Traits
4.4. Soil and Climate
4.5. Soil Microbial Community Composition
4.6. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Predictors | Variable | Acronym | Value Range | Units |
|---|---|---|---|---|
| Plant community composition | Plant richness | PSPRICH | 7–20 | number |
| Plant diversity | PDIV | 1.035–2.546 | Unitless | |
| Plant functional traits | Maximum plant height | Height | 2.09–4.59 | m |
| Specific leaf area | SLA | 0.07–0.39 | mm2 mg−1 | |
| Leaf dry matter content | LDMC | 97.35–298.07 | mg g−1 | |
| Leaf vein density | VD | 2.11–5.11 | mm mm−2 | |
| Leaf carbon content | LC | 164.90–454.54 | g kg−1 | |
| Leaf nitrogen content | LN | 11.38–30.67 | g kg−1 | |
| Leaf phosphorus content | LP | 0.71–2.16 | g kg−1 | |
| Root diameter | RD | 0.21–1.19 | mm | |
| Specific root length | SRL | 3.29–9.17 | m g−1 | |
| Root dry matter content | RDMC | 104.17–314.99 | mg g−1 | |
| Root carbon content | RC | 151.54–414.79 | g kg−1 | |
| Root nitrogen content | RN | 5.05–17.07 | g kg−1 | |
| Root phosphorus content | RP | 0.23–0.72 | g kg−1 | |
| Soil properties | Soil organic carbon | SOC | 12.64–61.37 | g kg−1 |
| Total nitrogen | TN | 0.9–4.01 | g kg−1 | |
| Total phosphorus | TP | 0.09–0.39 | g kg−1 | |
| Total potassium | TK | 8.07-41.02 | g kg−1 | |
| Soil pH | pH | 3.56–4.18 | Unitless | |
| Soil moisture | Sm | 16.7–48.34 | % | |
| C:N ratio | CN | 10.19–31.44 | Unitless |
| Plant Attributes | Sum of Squares | Mean of Squares | F | P |
|---|---|---|---|---|
| Plant richness | 36.22 | 12.07 | 1.45 | 0.25 |
| Plant diversity | 1.363 | 0.45 | 4.31 | 0.01 |
| Fungal richness | 207656.8 | 69218.92 | 9.91 | 0.01 |
| Fungal diversity | 20.45 | 6.82 | 6.31 | 0.01 |
| Bacteria richness | 6717.86 | 2239.29 | 7.98 | 0.01 |
| Bacteria diversity | 0.23 | 0.08 | 3.90 | 0.01 |
| Microbial Indices | Species Richness | Plant Diversity | CWM | FD | Forest Types | Model R2 (Adjusted) | Model Significance |
|---|---|---|---|---|---|---|---|
| Fungal richness | *** | NS | * | NS | *** | 0.69 | *** |
| Fungal diversity | NS | *** | ** | ** | *** | 0.67 | *** |
| Bacteria richness | ** | NS | NS | ** | * | 0.49 | *** |
| Bacteria diversity | NS | *** | NS | * | NS | 0.31 | ** |
| Species | Fungal Richness | Bacterial Richness | EMF | AMF | Pathogens |
|---|---|---|---|---|---|
| Illex asprella | 0.403 * | −0.117 | −0.313 | 0.454 ** | 0.091 |
| Clerodendrum fortunatum | −0.167 | 0.247 | 0.562 ** | −0.006 | −0.455 ** |
| Melicope pteleifolia | −0.077 | 0.337 * | 0.316 | −0.031 | −0.201 |
| Gardenia jasminoides | 0.101 | 0.409 * | 0.096 | 0.097 | −0.139 |
| Forests | Soil Organic Carbon (g kg−1) | Total Nitrogen (g kg−1) | Total Phosphorus (g kg−1) | Total Potassium (g kg−1) | pH | Soil Moisture (%) |
|---|---|---|---|---|---|---|
| AM | 44.067a | 2.4489a | 0.1894b | 16.479b | 3.7600b | 33.213 |
| EE | 27.234b | 1.4500b | 0.1397b | 9.951c | 3.7678b | 32.619 |
| MC | 28.600b | 1.6622b | 0.2978a | 34.666a | 3.9478a | 24.572 |
| NS | 23.918b | 1.3956b | 0.3525a | 26.755a | 3.9411a | 30.027 |
| P-value | 0.0007 | 0.0136 | 0.0000 | 0.0006 | 0.0062 | 0.0633 |
| Microbial Indices | SOC | TN | TP | TK | PH | SM | CN |
|---|---|---|---|---|---|---|---|
| Overall | |||||||
| Fungal richness | 0.717 ** | 0.674 ** | −0.254 | −0.197 | −0.218 | 0.04 | 0.3 |
| Fungal diversity | 0.566 ** | 0.421 * | −0.34 | −0.259 | −0.179 | 0.189 | 0.017 |
| Bacterial richness | −0.039 | −0.123 | −0.023 | −0.011 | −0.054 | −0.146 | 0.243 |
| Bacterial diversity | 0.373 * | 0.215 | −0.126 | −0.242 | −0.116 | 0.147 | −0.056 |
| Acacia | |||||||
| Fungal richness | 0.654 * | 0.268 | −0.175 | −0.014 | −0.495 | 0.521 | 0.443 |
| Fungal diversity | 0.357 | −0.151 | −0.635 | −0.644 | −0.308 | 0.211 | 0.424 |
| Bacterial richness | 0.721 * | 0.432 | −0.262 | −0.24 | −0.316 | 0.335 | 0.35 |
| Bacterial diversity | 0.767 * | 0.773 * | −0.553 | −0.381 | −0.665 | 0.119 | 0.13 |
| Eucalyptus | |||||||
| Fungal richness | 0.926 ** | 0.850 ** | 0.096 | 0.616 | 0.42 | −0.266 | −0.156 |
| Fungal diversity | −0.005 | 0.166 | −0.668 * | −0.242 | −0.146 | 0.214 | −0.26 |
| Bacterial richness | 0.665 * | 0.672 * | 0.199 | 0.069 | −0.051 | −0.226 | −0.247 |
| Bacterial diversity | 0.263 | 0.762 * | 0 | −0.175 | −0.038 | 0.412 | −0.862 ** |
| Coniferous | |||||||
| Fungal richness | 0.785 * | 0.5 | −0.045 | −0.840 ** | −0.299 | −0.048 | 0.435 |
| Fungal diversity | 0.419 | −0.068 | 0.594 | −0.333 | 0.462 | −0.115 | 0.422 |
| Bacterial richness | 0.733 * | 0.593 | −0.26 | −0.778 * | −0.277 | −0.178 | 0.315 |
| Bacterial diversity | 0.2 | 0.321 | 0.21 | −0.615 | −0.027 | 0.115 | −0.055 |
| Native | |||||||
| Fungal richness | 0.748 * | 0.292 | 0.388 | 0.493 | 0.176 | −0.096 | 0.666 |
| Fungal diversity | −0.094 | 0.246 | −0.41 | −0.063 | 0.236 | 0.222 | −0.22 |
| Bacterial richness | 0.687 * | 0.423 | −0.043 | 0.014 | −0.273 | −0.095 | 0.549 |
| Bacterial diversity | 0.277 | 0.716 * | 0.095 | 0.083 | 0.176 | 0.335 | 0.003 |
| Plant Traits | AM | EE | MC | NS | F | P |
|---|---|---|---|---|---|---|
| Single trait functional diversity | ||||||
| Height | 0.10 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.08 ± 0.01 | 1.09 | 0.3704 |
| SLA | 0.10 ± 0.01ab | 0.10 ± 0.01bc | 0.12 ± 0.01a | 0.08 ± 0.01c | 4.41 | 0.0132 |
| LDMC | 0.10 ± 0.01ab | 0.09 ± 0.01bc | 0.11 ± 0.01a | 0.08 ± 0.01c | 5.1 | 0.0072 |
| VD | 0.10 ± 0.01b | 0.10 ± 0.01b | 0.12 ± 0.01a | 0.08 ± 0.01c | 6.35 | 0.0025 |
| LC | 0.10 ± 0.01ab | 0.09 ± 0.01bc | 0.11 ± 0.01a | 0.07 ± 0.01c | 5.67 | 0.0044 |
| LN | 0.11 ± 0.01ab | 0.10 ± 0.01bc | 0.12 ± 0.01a | 0.08 ± 0.01c | 3.68 | 0.0261 |
| LP | 0.10 ± 0.01ab | 0.09 ± 0.01b | 0.12 ± 0.01a | 0.08 ± 0.01b | 3.39 | 0.0343 |
| RD | 0.10 ± 0.01ab | 0.10 ± 0.01b | 0.11 ± 0.01a | 0.08 ± 0.01b | 3.45 | 0.0325 |
| SRL | 0.24 ± 0.04a | 0.09 ± 0.01b | 0.13 ± 0.01b | 0.27 ± 0.04a | 4.25 | 0.0508 |
| RDMC | 0.28 ± 0.06a | 0.09 ± 0.01b | 0.13 ± 0.01b | 0.34 ± 0.06a | 7.67 | 0.0009 |
| RC | 0.28 ± 0.06a | 0.09 ± 0.01b | 0.13 ± 0.01b | 0.36 ± 0.06a | 8.28 | 0.0006 |
| RN | 0.29 ± 0.06a | 0.09 ± 0.01b | 0.13 ± 0.01b | 0.37 ± 0.06a | 7.66 | 0.0009 |
| RP | 0.28 ± 0.06a | 0.09 ± 0.01b | 0.13 ± 0.01b | 0.33 ± 0.06a | 7.42 | 0.0011 |
| Species | Family | Growth Form | Plant Type | Nitrogen Fixation |
|---|---|---|---|---|
| AM | ||||
| Acacia mangium | Fabaceae | Tree | Evergreen | Nitrogen fixing |
| Cinnamomum burmannii | Lauraceae | Tree | Evergreen | Non-nitrogen fixing |
| Eurya chinensis | Theaceae | Tree | Evergreen | Non-nitrogen fixing |
| Ilex asprella | Aquifoliaceae | Tree | Deciduous | Non-nitrogen fixing |
| Litsea cubeba | Lauraceae | Tree | Evergreen | Non-nitrogen fixing |
| Litsea rotundifolia | Lauraceae | Tree | Evergreen | Non-nitrogen fixing |
| Melicope pteleifolia | Rutaceae | Tree | Deciduous | Non-nitrogen fixing |
| Rhodomyrtus tomentosa | Myrtaceae | Tree | Evergreen | Non-nitrogen fixing |
| Clerodendrum fortunatum | Lamiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Gardenia jasminoides | Rubiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Litsea glutinosa | Lauraceae | Shrub | Evergreen | Non-nitrogen fixing |
| EE | ||||
| Eucalyptus exserta | Myrtaceae | Tree | Evergreen | Non-nitrogen fixing |
| Eucalyptus rostrata | Myrtaceae | Tree | Evergreen | Non-nitrogen fixing |
| Eucalyptus urophylla | Myrtaceae | Tree | Evergreen | Non-nitrogen fixing |
| Ilex asprella | Aquifoliaceae | Tree | Deciduous | Non-nitrogen fixing |
| Litsea cubeba | Lauraceae | Tree | Evergreen | Non-nitrogen fixing |
| Litsea glutinosa | Lauraceae | Tree | Evergreen | Non-nitrogen fixing |
| Melicope pteleifolia | Rutaceae | Tree | Deciduous | Non-nitrogen fixing |
| Clerodendrum fortunatum | Lamiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Gardenia jasminoides | Rubiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Melastoma malabathricum | Melastomataceae | Shrub | Evergreen | Non-nitrogen fixing |
| Rhodomyrtus tomentosa | Myrtaceae | Shrub | Evergreen | Non-nitrogen fixing |
| MC | ||||
| Cunninghamia lanceolata | Pinaceae | Tree | Evergreen | Non-nitrogen fixing |
| Ilex asprella | Aquifoliaceae | Tree | Deciduous | Non-nitrogen fixing |
| Melicop epteleifolia | Rutaceae | Tree | Deciduous | Non-nitrogen fixing |
| Pinus massoniana | Pinaceae | Tree | Evergreen | Non-nitrogen fixing |
| Schima superba | Theaceae | Tree | Evergreen | Non-nitrogen fixing |
| Rhodomyrtus tomentosa | Myrtaceae | Tree | Evergreen | Non-nitrogen fixing |
| Clerodendrum fortunatum | Lamiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Gardenia jasminoides | Rubiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Psychotria asiatica | Rubiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| NS | ||||
| Ilex asprella | Aquifoliaceae | Tree | Deciduous | Non-nitrogen fixing |
| Melicope pteleifolia | Rutaceae | Tree | Deciduous | Non-nitrogen fixing |
| Psychotria asiatica | Rubiaceae | Tree | Evergreen | Non-nitrogen fixing |
| Schima wallichii | Theaceae | Tree | Evergreen | Non-nitrogen fixing |
| Schima superba | Theaceae | Tree | Evergreen | Non-nitrogen fixing |
| Clerodendrum fortunatum | Lamiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Eurya chinensis | Theaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Gardenia jasminoides | Rubiaceae | Shrub | Evergreen | Non-nitrogen fixing |
| Rhodomyrtus tomentosa | Myrtaceae | Shrub | Evergreen | Non-nitrogen fixing |
References
- Guo, Y.; Hou, L.; Zhang, Z.; Zhang, J.; Cheng, J.; Wei, G.; Lin, Y. Soil microbial diversity during 30 years of grassland restoration on the loess plateau: Tight linkages with plant diversity. Land Degrad. Dev. 2019, 30, 1172–1182. [Google Scholar] [CrossRef]
- Wang, F.; Ding, Y.; Sayer, E.J.; Li, Q.; Zou, B.; Mo, Q.; Li, Y.; Lu, X.; Tang, J.; Zhu, W.; et al. Tropical forest restoration: Fast resilience of plant biomass contrasts with slow recovery of stable soil C stocks. Funct. Ecol. 2017, 31, 2344–2355. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Haack, S.E.; Lin, W.; Li, B.; Wu, L.; Fang, C.; Zhang, Z. Soil microbial community structure and metabolic activity of Pinus elliottii plantations across different stand ages in a subtropical area. PLoS ONE 2015, 10, e0135354. [Google Scholar] [CrossRef]
- Chen, F.; Zheng, H.; Zhang, K.; Ouyang, Z.; Lan, J.; Li, H.; Shi, Q. Changes in soil microbial community structure and metabolic activity following conversion from native Pinus massoniana plantations to exotic Eucalyptus plantations. For. Ecol. Manag. 2013, 291, 65–72. [Google Scholar] [CrossRef]
- Deng, J.; Yin, Y.; Zhu, W.; Zhou, Y. Variations in soil bacterial community diversity and structures among different revegetation types. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Dou, Y.; Huang, Y.; An, S. Links between soil fungal diversity and plant and soil properties on the Loess Plateau. Front. Microbiol. 2017, 8, 2198. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.E.; Eissenstat, D.M.; Hobbie, S.E.; Oleksyn, J.; Jagodzinski, A.M.; Reich, P.B.; Chadwick, O.A.; Chorover, J. Tree species effects on coupled cycles of carbon, nitrogen, and acidity in mineral soils at a common garden experiment. Biogeochemistry 2012, 111, 601–614. [Google Scholar] [CrossRef]
- You, Y.; Wang, J.; Huang, X.; Tang, Z.; Liu, S.; Sun, O.J. Relating microbial community structure to functioning in forest soil organic carbon transformation and turnover. Ecol. Evol. 2014, 4, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiang, W.; Wu, H.; Ouyang, S.; Zhou, B.; Zeng, Y.; Chen, Y.; Kuzyakov, Y. Tree species identity surpasses richness in affecting soil microbial richness and community composition in subtropical forests. Soil Biol. Biochem. 2019, 130, 113–121. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Y.; Fang, S.; Tian, Y. Tree species composition influenced microbial diversity and nitrogen availability in rhizosphere soil. Plant Soil Environ. 2015, 61, 438–443. [Google Scholar] [CrossRef]
- De Gannes, V.; Bekele, I.; Dipchansingh, D.; Wuddivira, M.N.; De Cairies, S.; Boman, M.; Hickey, W.J. Microbial community structure and function of soil following ecosystem conversion from native forests to teak plantation forests. Front. Microbiol. 2016, 7, 1976. [Google Scholar] [CrossRef]
- Zuo, X.; Wang, S.; Lv, P.; Zhou, X.; Zhao, X.; Zhang, T.; Zhang, J. Plant functional diversity enhances associations of soil fungal diversity with vegetation and soil in the restoration of semiarid sandy grassland. Ecol. Evol. 2016, 6, 318–328. [Google Scholar] [CrossRef]
- Tian, J.; He, N.; Hale, L.; Niu, S.; Yu, G.; Liu, Y.; Blagodatskaya, E.; Kuzyakov, Y.; Gao, Q.; Zhou, J. Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests. Funct. Ecol. 2017, 32, 61–70. [Google Scholar] [CrossRef]
- Zak, D.R.; Holmes, W.E.; White, D.C.; Peacock, A.D.; Tilman, D. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 2003, 84, 2042–2050. [Google Scholar] [CrossRef]
- Gould, I.J.; Quinton, J.N.; Weigelt, A.; De Deyn, G.B.; Bardgett, R.D. Plant diversity and root traits benefit physical properties key to soil function in grasslands. Ecol. Lett. 2016, 19, 1140–1149. [Google Scholar] [CrossRef]
- Petchey, O.; Gaston, K. Functional diversity: Back to basics and looking forward. Ecol. Lett. 2006, 9, 741–758. [Google Scholar] [CrossRef]
- LeBlanc, N.; Kinkel, L.L.; Kistler, H.C. Soil fungal communities respond to grassland plant community richness and soil edaphics. Microb. Ecol. 2015, 70, 188–195. [Google Scholar] [CrossRef]
- Sterkenburg, E.; Bahr, A.; BrandströmDurling, M.; Clemmensen, K.E.; Lindahl, B.D. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015, 207, 1145–1158. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Cajthaml, T.; Põlme, S.; Hiiesalu, I.; Anslan, S.; Harend, H.; Buegger, F.; Pritsch, K.; Koricheva, J.; et al. Tree diversity and species identity effects on soil fungi, protists and animals are context dependent. ISME J. 2016, 10, 346–362. [Google Scholar] [CrossRef]
- Roy-bolduc, A.; Lalibert, E.; Hijri, M. Strong linkage between plant and soil fungal communities along a successional coastal dune system. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef]
- Shi, L.; Mortimer, P.E.; Slik, J.W.F. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 2014, 64, 305–315. [Google Scholar] [CrossRef]
- Butterfield, B.J.; Suding, K.N. Single-trait functional indices outperform multi-trait indices in linking environmental gradients and ecosystem services in a complex landscape. J. Ecol. 2013, 101, 9–17. [Google Scholar] [CrossRef]
- Conti, G.; Díaz, S. Plant functional diversity and carbon storage–an empirical test in semi-arid forest ecosystems. J. Ecol. 2013, 101, 18–28. [Google Scholar] [CrossRef]
- Grime, J.P. Benefits of plant diversity to ecosystems: Immediate, filter and founder effects. J. Ecol. 1998, 86, 902–910. [Google Scholar] [CrossRef]
- de Vries, F.T.; Manning, P.; Tallowin, J.R.; Mortimer, S.R.; Pilgrim, E.S.; Harrison, K.A.; Hobbs, P.J.; Quirk, H.; Shipley, B.; Cornelissen, J.H.; et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 2012, 15, 1230–1239. [Google Scholar] [CrossRef]
- Tilman, D.; Reich, P.B.; Knops, J.; Wedin, D.; Mielke, T.; Lehman, C. Diversity and productivity in a long-term grassland experiment. Science 2001, 294, 843–845. [Google Scholar] [CrossRef]
- Legay, N.; Baxendale, C.; Grigulis, K.; Krainer, U.; Kastl, E.; Schloter, M.; Bardgett, R.D.; Arnoldi, C.; Bahn, M.; Dumont, N.; et al. Contribution of above-and below-ground plant traits to the structure and function of grassland soil microbial communities. Ann. Bot. 2014, 114, 1011–1021. [Google Scholar] [CrossRef]
- Grigulis, K.; Lavorel, S.; Krainer, U.; Legay, N.; Baxendal, C.; Dumont, M.; Kastl, E.; Arnoldi, C.; Bardgett, R.D.; Poly, F.; et al. Relative contributions of plant traits and soil microbial properties to mountain grassland ecosystem services. J. Ecol. 2013, 101, 47–57. [Google Scholar] [CrossRef]
- López-García, Á.; Varela-Cervero, S.; Vasar, M.; Öpik, M.; Barea, J.M.; Azcón-Aguilar, C. Plant traits determine the phylogenetic structure of arbuscular mycorrhizal fungal communities. Mol. Ecol. 2017, 26, 6948–6959. [Google Scholar] [CrossRef]
- Thion, C.E.; Poirel, J.D.; Cornulier, T.; De Vries, F.T.; Bardgett, R.D.; Prosser, J.I. Plant nitrogen-use strategy as a driver of rhizosphere archaeal and bacterial ammonia oxidiser abundance. FEMS Microbiol. Ecol. 2016, 92, 1–11. [Google Scholar] [CrossRef]
- De Long, J.R.; Jackson, B.G.; Wilkinson, A.; Pritchard, W.J.; Oakley, S.; Mason, K.E.; Stephan, J.G.; Ostle, N.J.; Johnson, D.; Baggs, E.M.; et al. Relationships between plant traits, soil properties and carbon fluxes differ between monocultures and mixed communities in temperate grassland. J. Ecol. 2019, 107, 1704–1719. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Fry, E.L.; Eldridge, D.J.; de Vries, F.T.; Manning, P.; Hamonts, K.; Kattge, J.; Boenisch, G.; Singh, B.K.; Bardgett, R.D. Plant attributes explain the distribution of soil microbial communities in two contrasting regions of the globe. New Phytol. 2018, 219, 574–587. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.; Keiblinger, K.M.; Schmid, E.; Sterflinger-Gleixner, K.; Ellersdorfer, G.; Roschitzki, B.; Richter, A.; Eberl, L.; Zechmeister-Boltenstern, S.; Riedel, K. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 2012, 6, 1749–1762. [Google Scholar] [CrossRef] [Green Version]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; Van Der Putten, W.H.; Wall, D.H. Ecological linkages between aboveground and belowground biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Shen, L.; Wen, C.; Yan, Q.; Ning, D.; Qin, Y.; Xue, K.; Wu, L.; He, Z.; et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 2016, 7, 12083. [Google Scholar] [CrossRef] [Green Version]
- Strickland, M.S.; Rousk, J. Considering fungal: Bacterial dominance in soils–methods, controls, and ecosystem implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Asuming-Brempong, S.; Nüsslein, K.; Marsh, T.L.; Flynn, S.J. Opening the black box of soil bacterial diversity. Appl. Soil Ecol. 1999, 13, 109–122. [Google Scholar] [CrossRef]
- Garnier, E.; Cortez, J.; Billès, G.; Navas, M.L.; Roumet, C.; Debussche, M.; Laurent, G.; Blanchard, A.; Aubry, D.; Bellmann, A.; et al. Plant functional markers capture ecosystem properties during secondary succession. Ecology 2004, 85, 2630–2637. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- Chen, Y.; Tian-Le, X.; Stavros, D.V.; Hang-Wei, H.; Zhi-Peng, H.; Ya-Jun, H.; Lei, L.; Ye, D.; Matthias, C.R.; Bao-Dong, C. Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biol. Biochem. 2017, 110, 12–21. [Google Scholar] [CrossRef]
- Hartman, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef]
- Sun, R.; Dsouza, M.; Gilbert, J.A.; Guo, X.; Wang, D.; Guo, Z.; Ni, Y.; Chu, H. Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ. Microbiol. 2016, 18, 5137–5150. [Google Scholar] [CrossRef]
- Yang, T.; Adams, J.M.; Shi, Y.; He, J.S.; Jing, X.; Chen, L.; Tedersoo, L.; Chu, H. Soil fungal diversity in natural grasslands of the Tibetan Plateau: Associations with plant diversity and productivity. New Phytol. 2017, 215, 756–765. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, Z.; Chen, C.; Jia, Z. Soil microbial community structure and diversity are largely influenced by soil pH and nutrient quality in 78-year-old tree plantations. Biogeosciences 2017, 14, 2101–2111. [Google Scholar] [CrossRef] [Green Version]
- Barberán, A.; McGuire, K.L.; Wolf, J.A.; Jones, F.A.; Wright, S.J.; Turner, B.L.; Essene, A.; Hubbell, S.P.; Faircloth, B.C.; Fierer, N. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecol. Lett. 2015, 18, 1397–1405. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Millard, P.; Singh, B.K. Does grassland vegetation drive soil microbial diversity? Nutr. Cycl. Agroecosystems 2010, 88, 147–158. [Google Scholar] [CrossRef]
- Navrátilová, D.; Tláskalová, P.; Kohout, P.; Dřevojan, P.; Fajmon, K.; Chytrý, M.; Baldrian, P. Diversity of fungi and bacteria in species-rich grasslands increases with plant diversity in shoots but not in roots and soil. FEMS Microb. Ecol. 2018, 95, fiy208. [Google Scholar] [CrossRef]
- Liu, J.; Dang, P.; Gao, Y.; Zhu, H.; Zhu, H.; Zhao, F.; Zhao, Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. For. Ecol. Manag. 2018, 427, 342–349. [Google Scholar] [CrossRef]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; Garcia-Gomez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantarel, A.A.; Pommier, T.; Desclos-Theveniau, M.; Diquélou, S.; Dumont, M.; Grassein, F.; Kastl, E.M.; Grigulis, K.; Laîné, P.; Lavorel, S.; et al. Using plant traits to explain plant–microbe relationships involved in nitrogen acquisition. Ecology 2015, 96, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Peìrez-Harguindeguy, N.; Díaz, S.; Garnier, E.; Lavorel, S.; Poorter, H.; Jaureguiberry, P. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013, 61, 167–234. [Google Scholar] [CrossRef]
- Wang, F.; Zhian, L.; Hanping, X.; Bi, Z.; Ningyu, L.; Jin, L.; Weixing, Z. Effects of nitrogen-fixing and non-nitrogen-fixing tree species on soil properties and nitrogen transformation during forest restoration in southern China. Soil Sci. Plant Nutr. 2010, 56, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Cornelissen, J.H.C.; Lavorel, S.; Garnier, E.; Diaz, S.; Buchmann, N.; Gurvich, D.E.; Reich, P.B.; Ter Steege, H.; Morgan, H.D.; Van Der Heijden, M.G.A.; et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 2003, 51, 335–380. [Google Scholar] [CrossRef] [Green Version]
- Gross, N.; Börger, L.; Soriano-Morales, S.I.; Le Bagousse-Pinguet, Y.; Quero, J.L.; García-Gómez, M.; Valencia-Gómez, E.; Maestre, F. Uncovering multiscale effects of aridity and biotic interactions on the functional structure of Mediterranean shrublands. J. Ecol. 2013, 101, 637–649. [Google Scholar] [CrossRef]
- Laliberté, E.; Legendre, P. A distance-based framework for measuring functional diversity from multiple traits. Ecology 2010, 91, 299–305. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.; Yan, Q.; Yang, S.; Van Nostrand, J.D.; Wang, Z.; He, Z.; Zhou, J.; Jiang, Y.; Deng, Y. Soil microbial beta-diversity is linked with compositional variation in aboveground plant biomass in a semi-arid grassland. Plant Soil 2018, 423, 465–480. [Google Scholar] [CrossRef]
- Han, Z.; Deng, M.; Yuan, A.; Wang, J.; Li, H.; Ma, J. Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 2018, 625, 106–113. [Google Scholar] [CrossRef]
- You, J.; Wu, G.; Ren, F.; Chang, Q.; Yu, B.; Xue, Y.; Mu, B. Microbial community dynamics in Baolige oilfield during MEOR treatment, revealed by Illumina MiSeq sequencing. Appl. Microbiol. Biotechnol. 2016, 100, 1469–1478. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F.; Smilauer, P. CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canoni-cal Community Ordination, Version 4.5. 2002. Available online: https://www.canoco.com and https://www.wur.nl/en/Publication-details.htm?publicationId=publication-way-333431383835 (accessed on 10 July 2018).
- Oksane, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package. R Package Version 2.3-0. 2015. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 14 March 2018).
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data, a comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]

| Plant Traits | AM | EE | MC | NS | F | P |
|---|---|---|---|---|---|---|
| Community weighted mean (CWM) trait | ||||||
| Height | 3.22±0.27 | 3.28 ± 0.23 | 3.14 ± 0.23 | 2.64 ± 0.15 | 1.66 | 0.2022 |
| SLA | 0.19 ± 0.01a | 0.15 ± 0.01b | 0.14 ± 0.01b | 0.14 ± 0.04b | 1.48 | 0.0212 |
| LDMC | 261.23 ± 11.23a | 197.85 ± 17.09b | 234.28 ± 6.01ab | 227.86 ± 6.10ab | 5.17 | 0.0067 |
| VD | 4.50 ± 0.28a | 3.98 ± 0.37ab | 4.09 ± 0.15a | 3.31 ± 0.16b | 3.83 | 0.0225 |
| LC | 386.66 ± 11.12 | 333.74 ± 29.26 | 370.49 ± 8.86 | 367.49 ± 11.59 | 1.53 | 0.2331 |
| LN | 20.87 ± 1.23a | 22.32 ± 1.95a | 21.60 ± 1.18a | 16.57 ± 1.25b | 3 | 0.0504 |
| LP | 1.75 ± 0.08a | 1.53 ± 0.13a | 1.24 ± 0.05b | 0.81 ± 0.04c | 23.16 | 0.0000 |
| RD | 0.51 ± 0.02 | 0.45 ± 0.04 | 0.48 ± 0.02 | 0.48 ± 0.09 | 0.25 | 0.8626 |
| SRL | 5.93 ± 0.25b | 6.56 ± 0.53ab | 5.81 ± 0.19b | 7.49 ± 0.20a | 6.1 | 0.0031 |
| RDMC | 262.00 ± 13.52a | 196.68 ± 16.97b | 224.92 ± 4.74b | 222.11 ± 5.39b | 6.19 | 0.0029 |
| RC | 378.89 ± 12.35a | 305.23 ± 26.62b | 354.30 ± 8.01a | 349 ± 11.37ab | 3.32 | 0.0367 |
| RN | 11.67 ± 0.39a | 11.20 ± 1.11a | 11.01 ± 0.36ab | 9.68 ± 0.28b | 1.75 | 0.0081 |
| RP | 0.56 ± 0.01 | 0.51 ± 0.04 | 0.58 ± 0.02 | 0.59 ± 0.02 | 1.59 | 0.2175 |
| Multi-trait Functional diversity | ||||||
| FD | 0.17 ± 0.02a | 0.09 ± 0.00b | 0.12 ± 0.00b | 0.17 ± 0.04a | 7.23 | 0.0013 |
| Soil Microbial Communities | Predictors | Indicators | % Variation Explained | P |
|---|---|---|---|---|
| Fungi | Taxonomic diversity Above ground traits Belowground traits | PSPRICH, Height, SLA, LDMC, VD, LN, LP, SRL, RDMC, RC, RN, RP | 0.76 | <0.001 |
| Above ground traits Belowground traits | Height, SLA, LDMC, VD, LN, LP, SRL, RDMC, RC, RN, RP | 0.68 | <0.001 | |
| Above ground traits | Height, LDMC, VD, LN, LP | 0.42 | <0.001 | |
| Belowground traits | SRL, RDMC, RC, RP | 0.43 | <0.001 | |
| Bacteria | Taxonomic diversity Above ground traits Belowground traits | PSPRICH, LDMC, VD, LC, LN, LP, RDMC, RN, RP | 0.45 | <0.001 |
| Above ground traits Belowground traits | LDMC, VD, LC, LN, LP, RDMC, RN, RP | 0.31 | <0.011 | |
| Above ground traits | LDMC, LC, LN, LP | 0.16 | <0.032 | |
| Belowground traits | RDMC, RC, RN, RP | 0.22 | <0.036 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanif, M.A.; Guo, Z.; Moniruzzaman, M.; He, D.; Yu, Q.; Rao, X.; Liu, S.; Tan, X.; Shen, W. Plant Taxonomic Diversity Better Explains Soil Fungal and Bacterial Diversity than Functional Diversity in Restored Forest Ecosystems. Plants 2019, 8, 479. https://doi.org/10.3390/plants8110479
Hanif MA, Guo Z, Moniruzzaman M, He D, Yu Q, Rao X, Liu S, Tan X, Shen W. Plant Taxonomic Diversity Better Explains Soil Fungal and Bacterial Diversity than Functional Diversity in Restored Forest Ecosystems. Plants. 2019; 8(11):479. https://doi.org/10.3390/plants8110479
Chicago/Turabian StyleHanif, Md. Abu, Zhiming Guo, M. Moniruzzaman, Dan He, Qingshui Yu, Xingquan Rao, Suping Liu, Xiangping Tan, and Weijun Shen. 2019. "Plant Taxonomic Diversity Better Explains Soil Fungal and Bacterial Diversity than Functional Diversity in Restored Forest Ecosystems" Plants 8, no. 11: 479. https://doi.org/10.3390/plants8110479





