Evolution of Target-Site Resistance to Glyphosate in an Amaranthus palmeri Population from Argentina and Its Expression at Different Plant Growth Temperatures
Abstract
:1. Introduction
2. Results
2.1. Resistance Confirmation Test
2.2. Mechanism of Resistance to Glyphosate
2.2.1. EPSPS Gene Sequencing
2.2.2. Level of Resistance Conferred by the P106S EPSPS Mutation and Other Potential Glyphosate Resistance Mechanisms in VM1
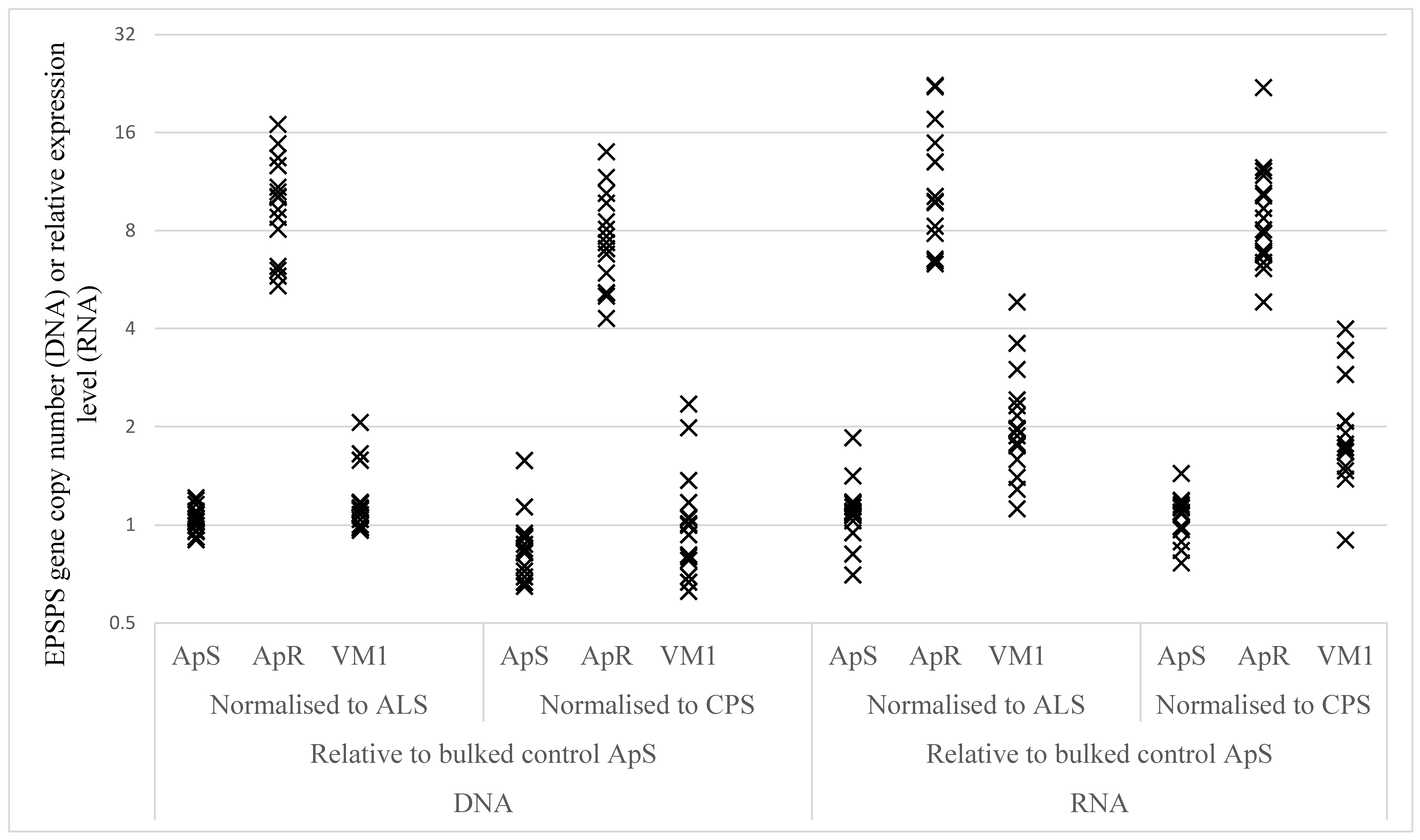
2.2.3. EPSPS Target Gene Duplication and Over-Expression
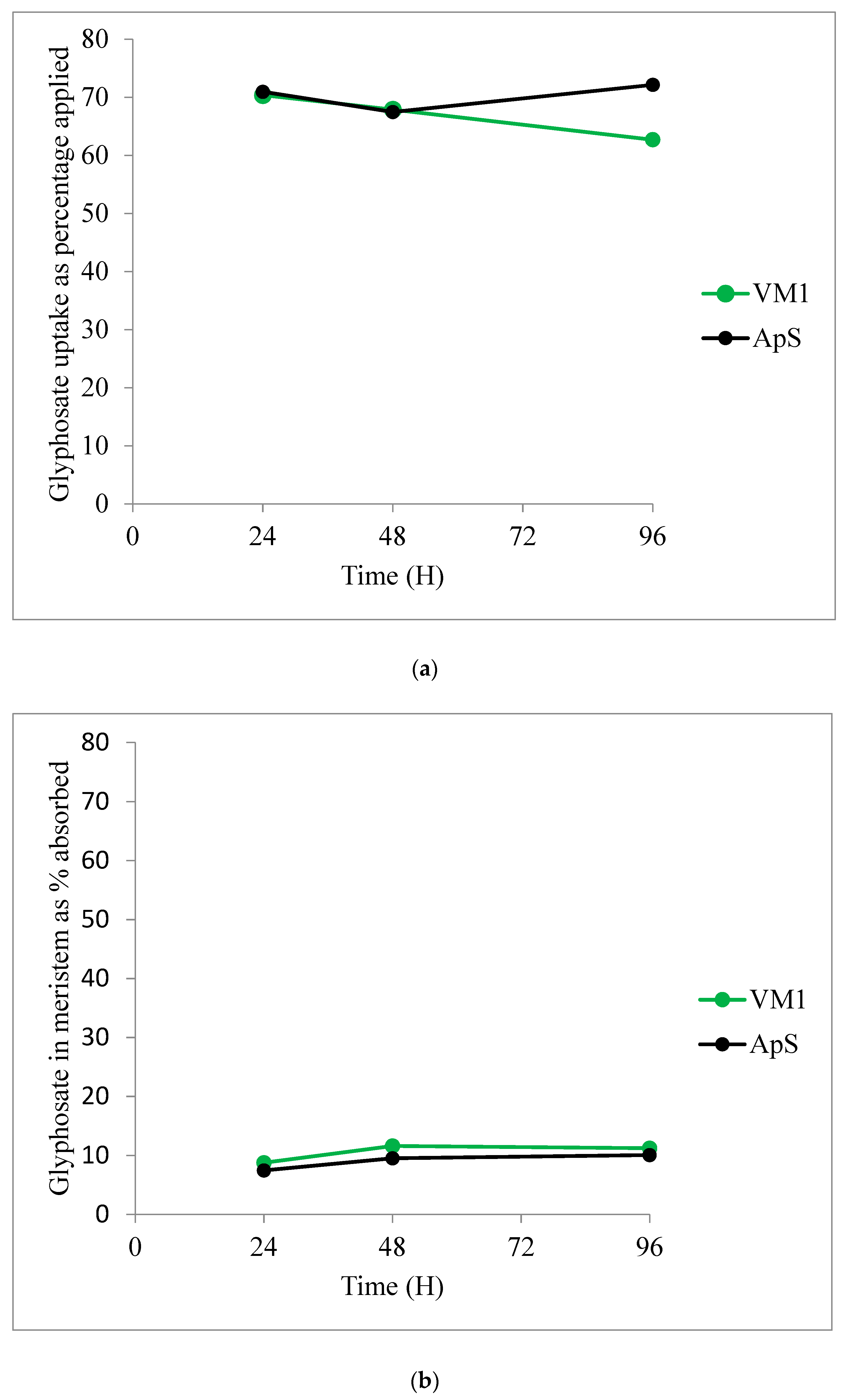
2.2.4. Glyphosate Uptake and Translocation
2.3. Relative Control of Wild and Mutant Plants at Low and High Temperatures
2.4. Prevalence of the P106S Mutation in 115 US A. palmeri Populations
3. Discussion
3.1. Mechanisms and Levels of Glyphosate Resistance Identified in VMI A. palmeri Population
3.2. Control of Wild and Mutant EPSPS Genotypes at Low and High Temperatures
3.3. Prevalence of the P106S Mutation in Native A. palmeri Populations
3.4. Implications for A. palmeri Management in Argentina
4. Materials and Methods
4.1. Materials
4.2. Plant Growth Conditions
4.3. Initial Glyphosate Resistance Confirmation Test
4.4. Mechanism of Resistance to Glyphosate
4.4.1. EPSPS Gene Sequencing Around Known Glyphosate-Resistance-Causing Mutations
4.4.2. Glyphosate Dose–Response Test on Precharacterised 106 EPSPS Genotypes
4.4.3. EPSPS Target Gene-Duplication and Over-Expression
4.4.4. Glyphosate Uptake and Translocation
4.5. Influence of Temperature on the Efficacy of Glyphosate on Wild and Mutant EPSPS Genotypes
4.6. Frequency of the P106S Mutation in a Large Number of A. palmeri from the USA
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sauer, J. Recent migration and evolution of the dioecious amaranths. Evolution 1957, 11, 11–31. [Google Scholar] [CrossRef]
- Bryson, C.; DeFelice, M. Weeds of the South; The University of Georgia Press: Athens, Greece, 2009; p. 34. [Google Scholar]
- Moerman, D. Native American Ethnobotany; Timber Press: Portland, OR, USA, 1998. [Google Scholar]
- Culpepper, A.S.; Webster, T.; Sosnoskie, L.; York, A. Glyphosate-Resistant Palmer Amaranth in the United States. In Glyphosate Resistance in Crops and Weeds: History, Development, and Management; Nandula, V.K., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 195–212. [Google Scholar]
- Ehleringer, J. Ecophysiology of Amaranthus palmeri, a sonoran desert summer annual. Oecologia 1983, 57, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.J.; Loughin, T.M. Growth analysis of four Amaranthus species. Weed Sci. 2000, 48, 347–355. [Google Scholar] [CrossRef]
- Keeley, P.; Carter, C.; Thullen, R. Influence of planting date on growth of palmer amaranth (Amaranthus palmeri). Weed Sci. 1987, 35, 199–204. [Google Scholar] [CrossRef]
- Steckel, L.E.; Sprague, C.L.; Stoller, E.W.; Wax, L.M. Temperature effects on germination of nine Amaranthus species. Weed Sci. 2004, 52, 217–221. [Google Scholar] [CrossRef]
- Forseth, I.N.; Ehleringer, J.R.; Werk, K.S.; Cook, C.S. Field water relations of Sonoran desert annuals. Ecology 1984, 65, 1436–1444. [Google Scholar] [CrossRef]
- Kilngaman, T.; Lawrence, R. Palmer amaranth (Amaranthus palmeri) interference in soybeans (Glycine max). Weed Sci. 1994, 42, 523–527. [Google Scholar] [CrossRef]
- Franssen, A.S.; Skinner, D.Z.; Al-Khatib, K.; Horak, M.J.; Kulakow, P.A. Interspecific hybridization and gene flow of ALS resistance in Amaranthus species. Weed Sci. 2001, 49, 598–606. [Google Scholar] [CrossRef]
- Chandi, A.; Milla-Lewis, S.R.; Jordan, D.L.; York, A.C.; Burton, J.D.; Zuleta, M.C.; Whitaker, J.R.; Culpepper, A.S. Use of AFLP markers to assess genetic diversity in Palmer amaranth (Amaranthus palmeri) populations from North Carolina and Georgia. Weed Sci. 2013, 61, 136–145. [Google Scholar] [CrossRef]
- Chahal, P.; Aulakh, J.; Jugulam, M.; Jhala, A. Herbicide-Resistant Palmer Amaranth (Amaranthus palmeri S. Wats.) in the United States—Mechanisms of Resistance, Impact, and Management. In Herbicides, Agronomic Crops and Weed Biology; Price, A., Kelton, J., Sarunaite, L., Eds.; InTech: Rijeka, Croatia, 2015; pp. 1–30. [Google Scholar]
- Gossett, B.; Murdock, E.; Toler, J. Resistance of Palmer amaranth (Amaranthus palmeri) to the dinitroaniline herbicides. Weed Technol. 1992, 6, 587–591. [Google Scholar] [CrossRef]
- Heap, I.M. International Survey of Herbicide-Resistant Weeds. Available online: http://www.weedscience.com (accessed on 15 November 2018).
- Kohrt, J.; Sprague, C.; Nadakuduti, S.S.; Douches, D. Confirmation of a three-way (glyphosate, ALS, and atrazine) herbicide-resistant population of Palmer amaranth (Amaranthus palmeri) in Michigan. Weed Sci. 2017, 65, 327–338. [Google Scholar] [CrossRef]
- Sosnoskie, L.M.; Webster, T.M.; Kichler, J.M.; MacRae, A.W.; Grey, T.L.; Culpepper, A.S. Pollen-mediated dispersal of glyphosate-resistance in Palmer amaranth under field conditions. Weed Sci. 2012, 60, 366–373. [Google Scholar] [CrossRef]
- Bagavathiannan, M.V.; Norsworthy, J.K. Multiple-Herbicide Resistance Is Widespread in Roadside Palmer Amaranth Populations. PLoS ONE 2016, 11, e0148748. [Google Scholar] [CrossRef] [PubMed]
- Rubione, C. Can herbicide resistance evolve due to factors other than a repeated use of technology? Argentina, a case to consider. Outlooks Pest Manag. 2017, 28, 213–219. [Google Scholar] [CrossRef]
- Hensleigh, P.; Pokorny, M. Palmer amaranth (Amaranthus palmeri S. Watson). In Agronomy Technical Note MT-92; USDA-NRCS: Washington, DC, USA, 2017. [Google Scholar]
- Matzrafi, M.; Herrmann, I.; Nansen, C.; Kliper, T.; Zait, Y.; Ignat, T.; Siso, D.; Rubin, B.; Karnieli, A.; Eizenberg, H. Hyperspectral technologies for assessing seed germination and trifloxysulfuron-methyl response in Amaranthus palmeri (Palmer amaranth). Front. Plant Sci. 2017, 8, 474. [Google Scholar] [CrossRef]
- Dominguez-Valenzuela, J.A.; Gherekhloo, J.; Fernández-Moreno, P.T.; Cruz-Hipolito, H.E.; Alcántara-de la Cruz, R.; Sánchez-González, E.; De Prado, R. First confirmation and characterization of target and non-target site resistance to glyphosate in Palmer amaranth (Amaranthus palmeri) from Mexico. Plant Physiol. Biochem. 2017, 115, 212–218. [Google Scholar] [CrossRef]
- Berger, S.; Madeira, P.; Ferrell, J.; Gettys, L.; Morichetti, S.; Cantero, J.; Nuñez, C. Palmer amaranth (Amaranthus palmeri) identification and documentation of ALS-resistance in Argentina. Weed Sci. 2016, 64, 312–320. [Google Scholar] [CrossRef]
- Küpper, A.; Borgato, E.A.; Patterson, E.L.; Gonçalves Netto, A.; Nicolai, M.; de Carvalho, S.J.P.; Nissen, S.J.; Gaines, T.A.; Christoffoleti, P.J. Multiple resistance to glyphosate and acetolactate synthase inhibitors in Palmer amaranth (Amaranthus palmeri) identified in Brazil. Weed Sci. 2017, 65, 317–326. [Google Scholar] [CrossRef]
- Morichetti, S.; Cantero, J.; Nunez, C.; Barboza, G.; Ariza-Espinar, L.; Amuchastegui, A.; Ferrell, J. Sobre la presencia de Amaranthus palmeri (Amaranthaceae) en Argentina. Bol. Soc. Argentian Bot. 2013, 48, 347–354. [Google Scholar]
- Heap, I.; Duke, S.O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 2017, 74, 1040–1049. [Google Scholar] [CrossRef]
- Tuesca, D.; Papa, J. Morichetti, Biology and management of Amaranthus palmeri in Argentina. In A Era Glyphosate: Agricultura, Meio-Ambiente e Homem; Meschede, D., Gazziero, D., Eds.; Midiograf: Londrina, Brazil, 2016; pp. 295–319. [Google Scholar]
- de Carvalho, L.B.; Alves, P.L.D.C.A.; González-Torralva, F.; Cruz-Hipolito, H.E.; Rojano-Delgado, A.M.; De Prado, R.; Gil-Humanes, J.; Barro, F.; Luque de Castro, M.D. Pool of resistance mechanisms to glyphosate in Digitaria insularis. J. Agric. Food Chem. 2012, 60, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Michitte, P.; De Prado, R.; Espinoza, N.; Ruiz-Santaella, J.; Gauvrit, C. Mechanisms of resistance to glyphosate in a ryegrass (Lolium multiflorum) biotype from Chile. Weed Sci. 2007, 55, 435–440. [Google Scholar] [CrossRef]
- Vila-Aiub, M.M.; Balbi, M.C.; Distéfano, A.J.; Fernández, L.; Hopp, E.; Yu, Q.; Powles, S.B. Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag. Sci. 2012, 68, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Brunharo, C.A.; Patterson, E.L.; Carrijo, D.R.; Melo, M.S.D.; Nicolai, M.; Gaines, T.A.; Nissen, S.J.; Christoffoleti, P.J. Confirmation and mechanism of glyphosate resistance in tall windmill grass (Chloris elata) from Brazil. Pest Manag. Sci. 2016, 72, 1758–1764. [Google Scholar] [CrossRef]
- Alcántara-de la Cruz, R.; Fernández-Moreno, P.T.; Ozuna, C.V.; Rojano-Delgado, A.M.; Cruz-Hipolito, H.E.; Domínguez-Valenzuela, J.A.; Barro, F.; De Prado, R. Target and non-target site mechanisms developed by glyphosate-resistant hairy beggarticks (Bidens pilosa L.) populations from Mexico. Front. Plant Sci. 2016, 7, 1492. [Google Scholar] [CrossRef]
- Moretti, M.; Hanson, B. Reduced translocation is involved in resistance to glyphosate and paraquat in Conyza bonariensis and Conyza canadensis from California. Weed Res. 2016, 57, 25–34. [Google Scholar] [CrossRef]
- Adu-Yeboah, P.; Malone, J.M.; Gill, G.; Preston, C. Reduced glyphosate translocation in two glyphosate-resistant populations of rigid ryegrass (Lolium rigidum) from fence lines in South Australia. Weed Sci. 2014, 62, 4–10. [Google Scholar] [CrossRef]
- Ganie, Z.A.; Jugulam, M.; Varanasi, V.K.; Jhala, A.J. Investigating the mechanism of glyphosate resistance in a common ragweed (Ambrosia artemisiifolia L.) biotype from Nebraska. Can. J. Plant Sci. 2017, 97, 1140–1151. [Google Scholar] [CrossRef]
- Nandula, V.K.; Wright, A.A.; Horn, C.R.V.; Molin, W.T.; Westra, P.; Reddy, K.N. Glyphosate resistance in giant ragweed (Ambrosia trifida L.) from Mississippi Is partly due to reduced translocation. Am. J. Plant Sci. 2015, 6, 2104–2113. [Google Scholar] [CrossRef]
- Gaines, T.; Zhang, W.; Wang, D.; Bukuna, B.; Chisholm, S.; Shaner, D.; Nissen, S.; Patzoldt, W.; Tranel, P.; Culpepper, A.; et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef]
- Molin, W.T.; Wright, A.A.; VanGessel, M.J.; McCloskey, W.B.; Jugulam, M.; Hoagland, R.E. Survey of the genomic landscape surrounding the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the USA. Pest Manag. Sci. 2018, 74, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Koo, D.-H.; Molin, W.T.; Saski, C.; Jiang, J.; Putta, K.; Jugulam, M.; Friebe, B.; Gill, B. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2018, 115, 3332–3337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohseni-Moghadam, M.; Schroeder, J.; Ashigh, J. Mechanism of resistance and inheritance in glyphosate resistant Palmer amaranth (Amaranthus palmeri) populations from New Mexico, USA. Weed Sci. 2013, 61, 517–525. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.; Lawton-Rauh, A.; Bagavathiannan, M.V.; Roma-Burgos, N. EPSPS gene amplification primarily confers glyphosate resistance among Arkansas Palmer amaranth (Amaranthus palmeri) populations. Weed Sci. 2018, 66, 293–300. [Google Scholar] [CrossRef]
- Lorentz, L.; Gaines, T.A.; Nissen, S.J.; Westra, P.; Strek, H.J.; Dehne, H.W.; Ruiz-Santaella, J.P.; Beffa, R. Characterization of glyphosate resistance in Amaranthus tuberculatus populations. J. Agric. Food Chem. 2014, 62, 8134–8142. [Google Scholar] [CrossRef]
- Baerson, S.R.; Rodriguez, D.J.; Biest, N.A.; Tran, M.; You, J.; Kreuger, R.W.; Dill, G.M.; Pratley, J.E.; Gruys, K.J. Investigating the mechanism of glyphosate resistance in rigid ryegrass (Lolium ridigum). Weed Sci. 2002, 50, 721–730. [Google Scholar] [CrossRef]
- Dinelli, G.; Marotti, I.; Bonetti, A.; Minelli, M.; Catizone, P.; Barnes, J. Physiological and molecular insight on the mechanisms of resistance to glyphosate in Conyza canadensis (L.) Cronq. biotypes. Pestic. Biochem. Physiol. 2006, 86, 30–41. [Google Scholar] [CrossRef]
- Baerson, S.R.; Rodriguez, D.J.; Tran, M.; Feng, Y.; Biest, N.A.; Dill, G.M. Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol. 2002, 129, 1265–1275. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Jalaludin, A.; Han, H.; Chen, M.; Sammons, R.D.; Powles, S.B. Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 2015, 167, 1440–1447. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Yu, Q.; Widderick, M.J.; Powles, S.B. Target-site EPSPS Pro-106 mutations: Sufficient to endow glyphosate resistance in polyploid Echinochloa colona? Pest Manag. Sci. 2016, 72, 264–271. [Google Scholar] [CrossRef]
- Page, E.R.; Grainger, C.M.; Laforest, M.; Nurse, R.E.; Rajcan, I.; Bae, J.; Tardif, F.J. Target and non-target site mechanisms confer resistance to glyphosate in Canadian accessions of Conyza canadensis. Weed Sci. 2018, 66, 234–245. [Google Scholar] [CrossRef]
- Bell, M.S.; Hager, A.G.; Tranel, P.J. Multiple resistance to herbicides from four site-of-action groups in waterhemp (Amaranthus tuberculatus). Weed Sci. 2013, 61, 460–468. [Google Scholar] [CrossRef]
- Ngo, T.D.; Krishnan, M.; Boutsalis, P.; Gill, G.; Preston, C. Target-site mutations conferring resistance to glyphosate in feathertop Rhodes grass (Chloris virgata) populations in Australia. Pest Manag. Sci. 2018, 74, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Simarmata, M.; Penner, D. The basis for glyphosate resistance in rigid Ryegrass (Lolium rigidum) from California. Weed Sci. 2008, 56, 181–188. [Google Scholar] [CrossRef]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.S.; Kabsch, W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef] [Green Version]
- Healy-Fried, M.L.; Funke, T.; Priestman, M.A.; Han, H.; Schönbrunn, E. Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J. Biol. Chem. 2007, 282, 32949–32955. [Google Scholar] [CrossRef] [Green Version]
- Sammons, R.; Gaines, T. Glyphosate resistance: State of knowledge. Pest Manag. Sci. 2014, 70, 1367–1377. [Google Scholar] [CrossRef] [Green Version]
- Kaundun, S.S.; Zelaya, I.; Dale, R.; Lycett, A.; Carter, P.; Sharples, P.; McIndoe, E. Importance of the P106S target site mutation in conferring resistance to glyphosate in an Eleusine indica biotype from the Philippines. Weed Sci. 2008, 56, 637–646. [Google Scholar] [CrossRef]
- Janel, L.H.; Riggins, C.W.; Steckel, L.E.; Tranel, P.J. The EPSPS Pro106Ser substitution solely accounts for glyphosate resistance in a goosegrass (Eleusine indica) population from Tennessee, United States. J. Integr. Agric. 2016, 15, 1304–1312. [Google Scholar] [CrossRef] [Green Version]
- Alarcón-Reverte, R.; García, A.; Watson, S.B.; Abdallah, I.; Sabaté, S.; Hernández, M.J.; Dayan, F.E.; Fischer, A.J. Concerted action of target-site mutations and high EPSPS activity in glyphosate-resistant junglerice (Echinochloa colona) from California. Pest Manag. Sci. 2015, 71, 996–1007. [Google Scholar] [CrossRef]
- Dennis, M.; Hembree, K.J.; Bushoven, J.T.; Shrestha, A. Growth stage, temperature, and time of year affects the control of glyphosate-resistant and glyphosate-paraquat resistant Conyza bonariensis with saflufenacil. Crop Prot. 2016, 81, 129–137. [Google Scholar] [CrossRef]
- Godar, A.S.; Varanasi, V.K.; Nakka, S.; Prasad, P.V.V.; Thompson, C.R.; Mithila, J. Physiological and molecular mechanisms of differential sensitivity of Palmer Amaranth (Amaranthus palmeri) to mesotrione at varying growth temperatures. PLoS ONE 2015, 10, e0126731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzrafi, M.; Seiwert, B.; Reemtsma, T.; Rubin, B.; Peleg, Z. Climate change increases the risk of herbicide-resistant weeds due to enhanced detoxification. Planta 2016, 244, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, C.G.; Jordan, T.N.; Wills, G.D. Translocation of 14C-glyphosate in soybeans (Glycine max) and Johnsongrass (Sorghum halepense). Weed Sci. 1980, 28, 113–118. [Google Scholar] [CrossRef]
- Vila-Aiub, M.; Gundel, P.E.; Yu, Q.; Powles, S.B. Glyphosate resistance in Sorghum halepense and Lolium rigidum is reduced at suboptimal growing temperatures. Pest Manag. Sci. 2013, 69, 228–232. [Google Scholar] [CrossRef]
- Ganie, Z.A.; Jugulam, M.; Jhala, A.J. Temperature influences efficacy, absorption, and translocation of 2,4-D or glyphosate in glyphosate-resistant and glyphosate-susceptible common ragweed (Ambrosia artemisiifolia) and giant ragweed (Ambrosia trifida). Weed Sci. 2017, 65, 588–602. [Google Scholar] [CrossRef]
- Pline, W.A.; Wu, J.; Hatzios, K.K. Effects of temperature and chemical additives on the response of transgenic herbicide-resistant soybeans to glufosinate and glyphosate applications. Pestic. Biochem. Physiol. 1999, 65, 119–131. [Google Scholar] [CrossRef]
- Ou, J.; Stahlman, P.; Jugulam, M. Reduced absorption of glyphosate anddecreased translocation of dicamba contributeto poor control of kochia (Kochia scoparia) at high temperature. Pest Manag. Sci. 2018, 74, 1134–1142. [Google Scholar] [CrossRef]
- Kleinman, Z.; Ben-Ami, G.; Rubin, B. From sensitivity to resistance—Factors affecting the response of Conyza spp. to glyphosate. Pest Manag. Sci. 2016, 72, 1681–1688. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Malone, J.M.; Boutsalis, P.; Shirley, N.; Preston, C. Temperature influences the level of glyphosate resistance in barnyardgrass (Echinochloa colona). Pest Manag. Sci. 2016, 72, 1031–1039. [Google Scholar] [CrossRef]
- Ge, X.; d’Avignon, D.A.; Ackerman, J.J.; Duncan, B.; Spaur, M.B.; Sammons, R.D. Glyphosate-resistant horseweed made sensitive to glyphosate: Low-temperature suppression of glyphosate vacuolar sequestration revealed by 31P NMR. Pest Manag. Sci. 2011, 67, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Shaner, D.L. Testing Methods for Glyphosate Resistance. In Glyphosate Resistance in Crops and Weeds: History, Development, and Management; Nandula, V.K., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 93–118. [Google Scholar]
- Burgos, N.R.; Tranel, P.J.; Streibig, J.C.; Davis, V.M.; Shaner, D.; Norsworthy, J.K.; Ritz, C. Review: Confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci. 2013, 61, 4–20. [Google Scholar] [CrossRef]
- Rios, S.I.; Wright, S.D.; Banuelos, G.; Shrestha, A. Tolerance of Amaranthus palmeri populations from California to postemergence herbicides at various growth stages. Crop Prot. 2016, 87, 6–12. [Google Scholar] [CrossRef]
- Vila-Aiub, M.; Casas, C.; Gundel, P.E. The role of plant size in the selection of glyphosate resistance in Sorghum halepense. Pest Manag. Sci. 2018, 74, 2464–2467. [Google Scholar] [CrossRef] [PubMed]
- Crow, W.; Steckel, L.; Mueller, T.; Hayes, R. Management of large, glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in corn. Weed Technol. 2016, 30, 611–616. [Google Scholar] [CrossRef]
- Gaines, T.A.; Shaner, D.L.; Ward, S.M.; Leach, J.E.; Preston, C.; Westra, P. Mechanism of resistance of evolved glyphosate-resistant Palmer amaranth (Amaranthus palmeri). J. Agric. Food Chem. 2011, 59, 5886–5889. [Google Scholar] [CrossRef]
- Sutton, M.C.; Klein, N.; Taylor, G. A comparative analysis of soybean production between the United States, Brazil, and Argentina. J. ASFMRA 2005, 9, 33–41. [Google Scholar]
- Ward, S.; Webster, T.; Steckel, L. Palmer amaranth (Amaranthus palmeri): A review. Weed Technol. 2013, 27, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Vivian, R.; Reis, A.; Kálnay, P.A.; Vargas, L.; Ferreira, A.C.C.; Mariani, F. Weed Management in Soybean—Issues and Practices. In Soybean—Pest Resistance; El-Shemy, H., Ed.; In Tech: Rijeka, Croatia, 2013; pp. 47–84. [Google Scholar]
- Liu, C.; Bridges, M.E.; Kaundun, S.S.; Glasgow, L.; Owen, M.D.K.; Neve, P. A generalised individual-based algorithm for modelling the evolution of quantitative herbicide resistance in arable weed populations. Pest Manag. Sci. 2017, 73, 462–474. [Google Scholar] [CrossRef]

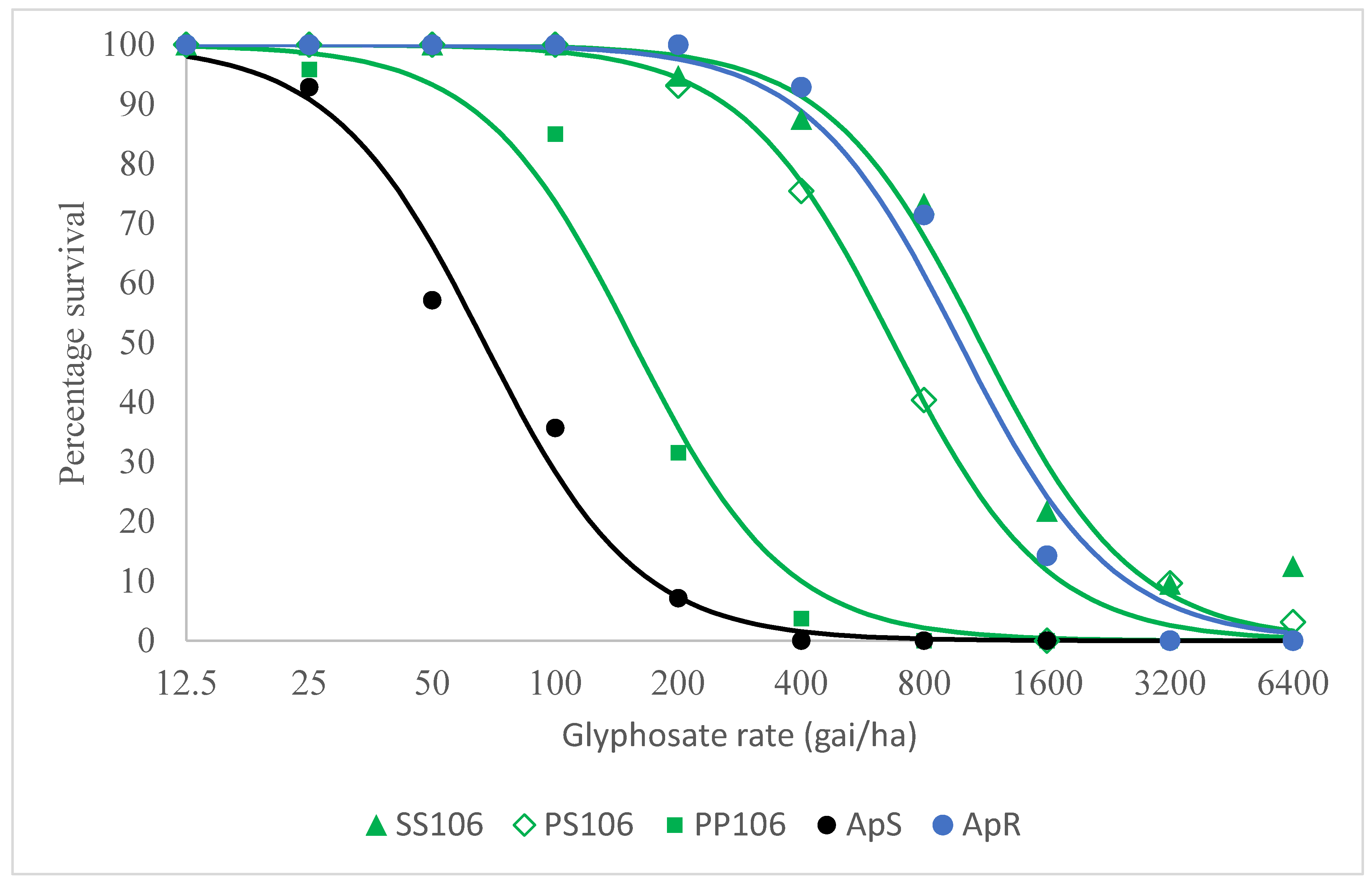

| (a) | |
| Group | LD50 |
| SS106 | 1102 (857–1414) |
| PS106 | 672 (578–783) |
| PP106 | 155 (123–196) |
| ApS | 67 (50–90) |
| ApR | 977 (732–1305) |
| (b) | |
| Comparison | Resistance Index |
| SS106 vs PP106 | 7.1 (5.0–10.0) |
| PS106 vs PP106 | 4.3 (3.3–5.7) |
| PP106 vs ApS | 2.3 (1.6–3.4) |
| SS106 vs ApS | 16.4 (11.2–24.1) |
| PS106 vs ApS | 10.0 (7.2–13.9) |
| ApR vs ApS | 14.6 (9.7–22.0) |
| Sample | Gene Comparison | ApS | ApR | VM1 | ApR vs. ApS | VM1 vs. ApS | ||
|---|---|---|---|---|---|---|---|---|
| Ratio | P-Value | Ratio | P-Value | |||||
| DNA | EPSPS vs CPS | 0.85 | 7.35 | 0.99 | 8.68 | <0.0001 | 1.17 | 0.1553 |
| DNA | EPSPS vs ALS | 1.04 | 9.43 | 1.18 | 9.07 | <0.0001 | 1.13 | 0.1450 |
| RNA | EPSPS vs CPS | 1.05 | 8.81 | 1.87 | 8.37 | <0.0001 | 1.78 | <0.0001 |
| RNA | EPSPS vs ALS | 1.10 | 10.85 | 2.04 | 9.90 | <0.0001 | 1.86 | <0.0001 |
| Time after Treatment | 24 h | 48 h | 72 h | |||
|---|---|---|---|---|---|---|
| Population | ApS | VM1 | ApS | VM1 | ApS | VM1 |
| Treated leaf | 73.4 ± 1.8 | 68.2 ± 4.9 | 74.9 ± 7.4 | 62.7 ± 7.7 | 71.9 ± 10.3 | 39.7 ± 6.0 |
| Meristem | 7.5 ± 0.7 | 8.8 ± 1.2 | 9.5 ± 2.3 | 11.6 ± 2.7 | 10.1 ± 3.7 | 11.2 ± 1.1 |
| Rest of plant | 19.1 ± 2.0 | 23.0 ± 3.8 | 15.5 ± 5.3 | 25.7 ± 5.2 | 18.0 ± 6.6 | 49.1 ± 6.6 |
| (a) | |||||
| Growth Condition | Test | PP106-ApS | PP106-VM1-P | PS106-VM1-P | SS106-VM1-P |
| 20 °C day/16 °C night | 1 | 0/20 = 0% | 0/43 = 0% | 15/91 = 16.5% | 9/34 = 26.5% |
| 2 | 0/28 = 0% | 4/31 = 12.9% | 35/81 = 43.2% | 34/58 = 58.6% | |
| 30 °C day/26 °C night | 1 | 0/20 = 0% | 4/35 = 11.4% | 61/91 = 67.0% | 32/43 = 74.4% |
| 2 | 0/28 = 0% | 9/40 = 22.5% | 73/92 = 79.3% | 34/38 = 89.5% | |
| (b) | |||||
| 20 °C day/16 °C Night | 30 °C day/26 °C Night | ||||
| Comparison | CMH statistic | P-value | CMH statistic | P-value | |
| PS106-VM1-P vs. PP106-VM1-P | 16.7 | <0.0001 | 68.9 | <0.0001 | |
| SS106-VM1-P vs. PP106-VM1-P | 29.1 | <0.0001 | 65.2 | <0.0001 | |
| SS106-VM1-P vs. PS106-VM1-P | 4.8 | 0.0291 | 2.3 | 0.1264 | |
| PP106-VM1-P vs. PP106-ApS | 3.8 | 0.0509 | 9.5 | 0.0020 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaundun, S.S.; Jackson, L.V.; Hutchings, S.-J.; Galloway, J.; Marchegiani, E.; Howell, A.; Carlin, R.; Mcindoe, E.; Tuesca, D.; Moreno, R. Evolution of Target-Site Resistance to Glyphosate in an Amaranthus palmeri Population from Argentina and Its Expression at Different Plant Growth Temperatures. Plants 2019, 8, 512. https://doi.org/10.3390/plants8110512
Kaundun SS, Jackson LV, Hutchings S-J, Galloway J, Marchegiani E, Howell A, Carlin R, Mcindoe E, Tuesca D, Moreno R. Evolution of Target-Site Resistance to Glyphosate in an Amaranthus palmeri Population from Argentina and Its Expression at Different Plant Growth Temperatures. Plants. 2019; 8(11):512. https://doi.org/10.3390/plants8110512
Chicago/Turabian StyleKaundun, Shiv Shankhar, Lucy Victoria Jackson, Sarah-Jane Hutchings, Jonathan Galloway, Elisabetta Marchegiani, Anushka Howell, Ryan Carlin, Eddie Mcindoe, Daniel Tuesca, and Raul Moreno. 2019. "Evolution of Target-Site Resistance to Glyphosate in an Amaranthus palmeri Population from Argentina and Its Expression at Different Plant Growth Temperatures" Plants 8, no. 11: 512. https://doi.org/10.3390/plants8110512
APA StyleKaundun, S. S., Jackson, L. V., Hutchings, S.-J., Galloway, J., Marchegiani, E., Howell, A., Carlin, R., Mcindoe, E., Tuesca, D., & Moreno, R. (2019). Evolution of Target-Site Resistance to Glyphosate in an Amaranthus palmeri Population from Argentina and Its Expression at Different Plant Growth Temperatures. Plants, 8(11), 512. https://doi.org/10.3390/plants8110512





