Characterization of Cover Crop Rooting Types from Integration of Rhizobox Imaging and Root Atlas Information
Abstract
:1. Introduction
2. Results
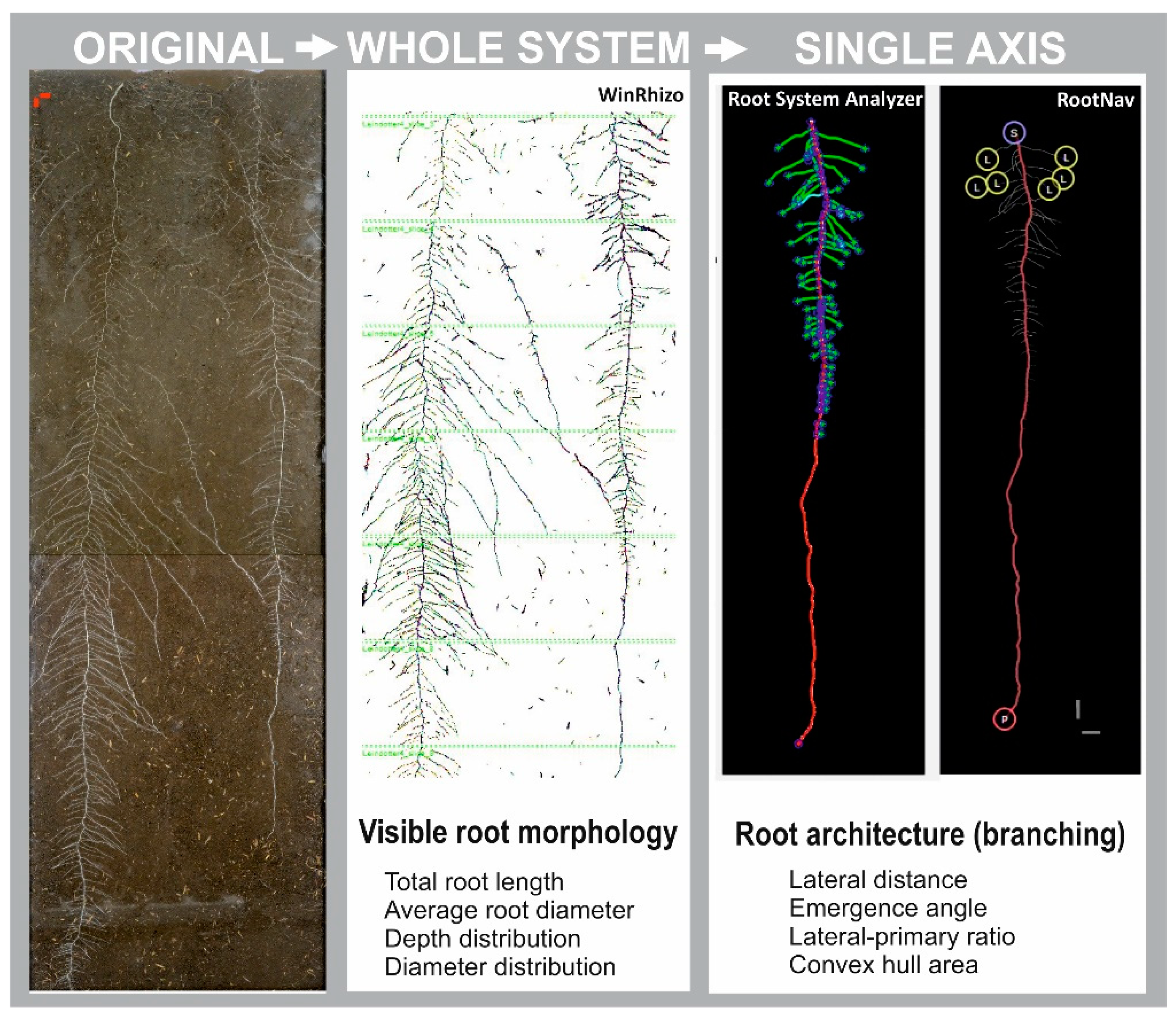
2.1. Rhizobox-Based Root System Characterization
2.2. Root System Characterization from Root Atlas Data
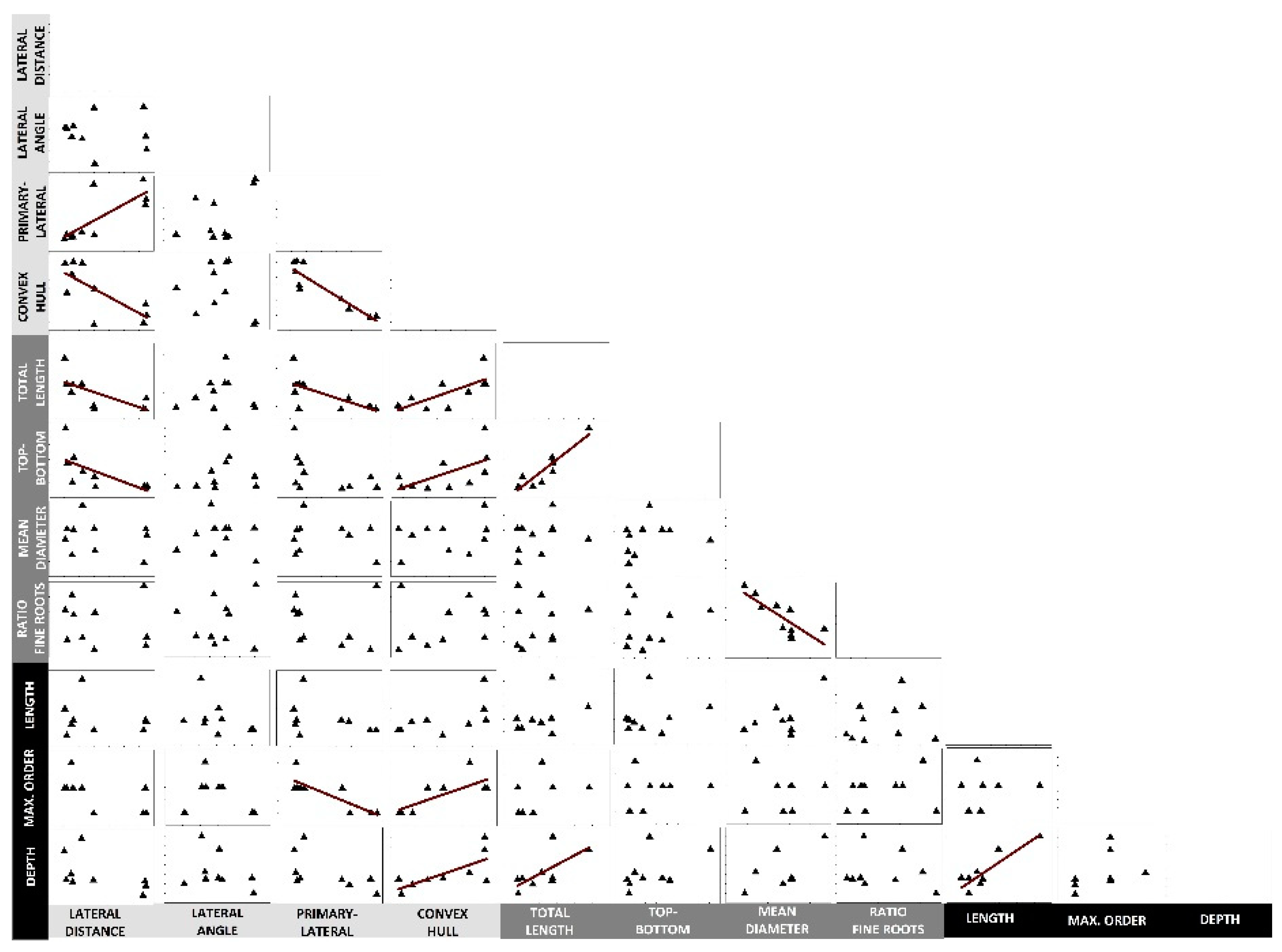
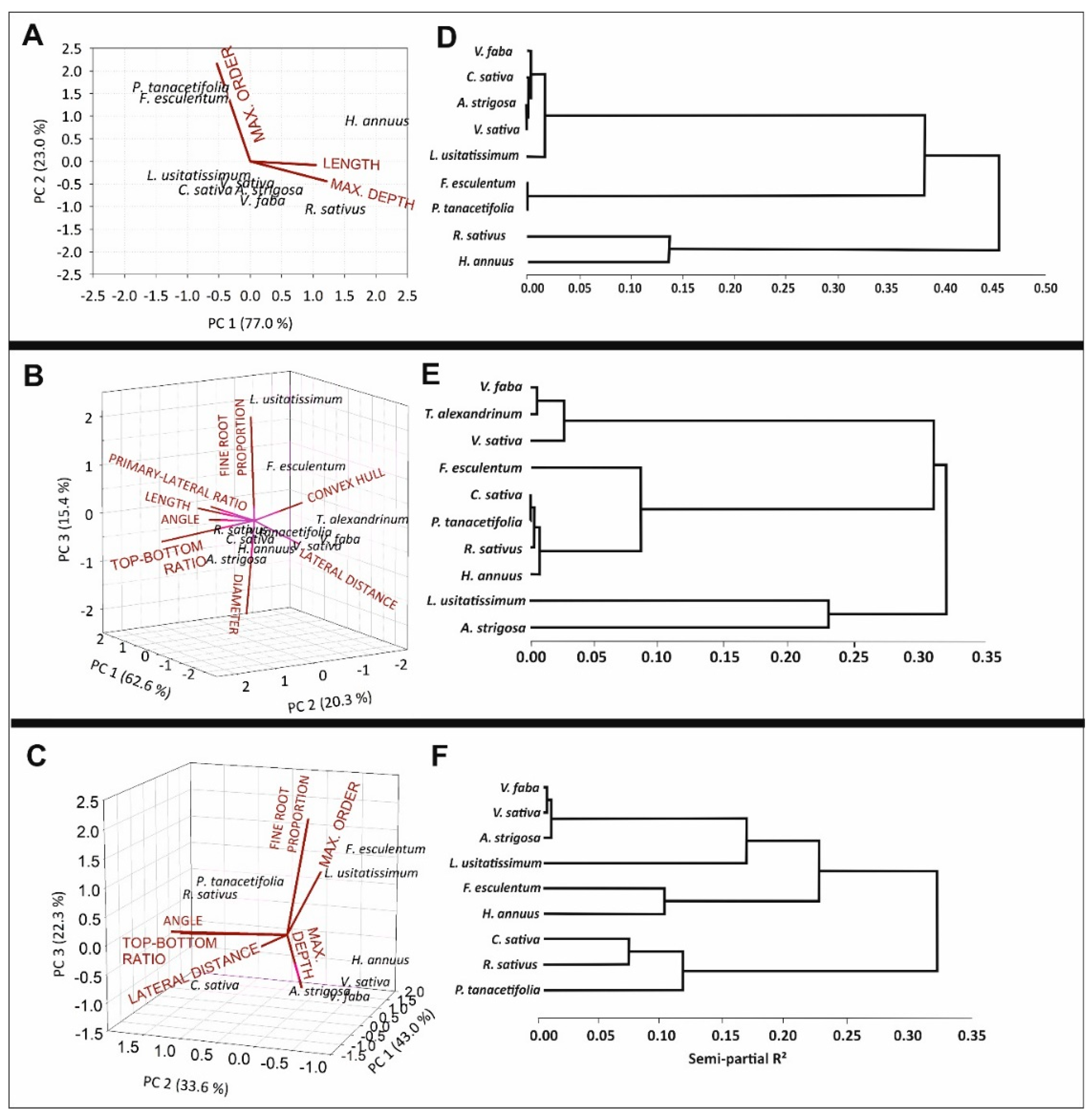
2.3. Inter-Trait Relations
2.4. Trait-Based Root Classification of Species
3. Discussion
3.1. Root Data Integration
3.2. Root System Descriptors for Comprehensive Species Characterization
3.3. Cover Crop Rooting Types
4. Materials and Methods
4.1. Plant Material
4.2. Rhizobox Experiment
4.2.1. Experimental Setup and Growing Conditions
4.2.2. Imaging and Image Analysis
4.3. Root Atlas Data
4.4. Statistical Evaluation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kenrick, P.; Strullu-Derrien, C. The Origin and Early Evolution of Roots. Plant Physiol. 2014, 166, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Goebel, K. Organographie der Pflanzen; III Aufl. II Teil. G. Fischer: Jena, Germany, 1930. [Google Scholar]
- Zobel, R.W.; Waisel, Y. A plant root system architectural taxonomy: A framework for root nomenclature. Plant Biosyst. 2010, 144, 507–512. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Woll, K.; Sauer, M.; Dembinsky, D. Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann. Bot. 2004, 93, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Fitter, A.H. An architectural approach to the comparative ecology of plant root systems. New Phytol. 1987, 106, 61–77. [Google Scholar] [CrossRef]
- Taub, D.R.; Goldberg, D. Root system topology of plants from habitats differing in soil resource availability. Funct. Ecol. 1996, 10, 258–264. [Google Scholar] [CrossRef]
- Cruz, C.; Green, J.J.; Watson, C.A.; Wilson, F.; Martins-Loução, M.A. Functional aspects of root architecture and mycorrhizal inoculation with respect to nutrient uptake capacity. Mycorrhiza 2004, 14, 177–184. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, D.; Xu, X.; Lu, M.; Bardgett, R.D.; Eissenstat, D.M.; McCormack, M.L.; Hedin, L.O. Evolutionary history resolves global organization of root functional traits. Nature 2018, 555, 94–97. [Google Scholar] [CrossRef]
- Lynch, J.P. Rhizoeconomics: The roots of shoot growth limitations. HortScience 2007, 42, 1107–1109. [Google Scholar] [CrossRef]
- Weaver, J.E.; Bruner, W.E. Root Development of Field Crops; McGraw-Hill Book Company: New York, NY, USA, 1926. [Google Scholar]
- Smit, A.L.; Bengough, A.G.; Engels, C.; van Noordwijk, M.; Pellerin, S.; van de Geijn, S.C. Root Methods: A Handbook; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Kuijken, R.C.P.; van Eeuwijk, F.A.; Marcelis, L.F.M.; Bouwmeester, H.J. Root phenotyping: From component trait in the lab to breeding. J. Exp. Bot. 2015, 66, 5389–5401. [Google Scholar] [CrossRef]
- Nagel, K.A.; Putz, A.; Gilmer, F.; Heinz, K.; Fischbach, A.; Pfeifer, J.; Faget, M.; Blossfeld, S.; Ernst, M.; Dimaki, C. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct. Plant Biol. 2012, 39, 891–904. [Google Scholar] [CrossRef]
- Bodner, G.; Nakhforoosh, A.; Arnold, T.; Leitner, D. Hyperspectral imaging: A novel approach for plant root phenotyping. Plant Methods 2018, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Metzner, R.; Eggert, A.; van Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Methods 2015, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Pound, M.P.; Bennett, M.J.; Wells, D.M. Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 2019, 55, 1–8. [Google Scholar] [CrossRef]
- Paez-Garcia, A.; Motes, C.; Scheible, W.-R.; Chen, R.; Blancaflor, E.; Monteros, M. Root traits and phenotyping strategies for plant improvement. Plants 2015, 4, 334–355. [Google Scholar] [CrossRef]
- Krajewski, P.; Chen, D.; Ćwiek, H.; van Dijk, A.D.J.; Fiorani, F.; Kersey, P.; Klukas, C.; Lange, M.; Markiewicz, A.; Nap, J.P. Towards recommendations for metadata and data handling in plant phenotyping. J. Exp. Bot. 2015, 66, 5417–5427. [Google Scholar] [CrossRef]
- Tardieu, F. Any trait or trait-related allele can confer drought tolerance: Just design the right drought scenario. J. Exp. Bot. 2011, 63, 25–31. [Google Scholar] [CrossRef]
- Wezel, A.; Casagrande, M.; Celette, F.; Vian, J.-F.; Ferrer, A.; Peigné, J. Agroecological practices for sustainable agriculture. A review. Agron. Sustain. Dev. 2014, 34, 1–20. [Google Scholar] [CrossRef]
- Kaye, J.P.; Quemada, M. Using cover crops to mitigate and adapt to climate change. A review. Agron. Sustain. Dev. 2017, 37, 4. [Google Scholar] [CrossRef]
- Wittwer, R.A.; Dorn, B.; Jossi, W.; Van Der Heijden, M.G.A. Cover crops support ecological intensification of arable cropping systems. Sci. Rep. 2017, 7, 41911. [Google Scholar] [CrossRef]
- Kristensen, H.L.; Thorup-Kristensen, K. Root growth and nitrate uptake of three different catch crops in deep soil layers. Soil Sci. Soc. Am. J. 2004, 68, 529–537. [Google Scholar] [CrossRef]
- Yu, Y.; Loiskandl, W.; Kaul, H.-P.; Himmelbauer, M.; Wie, W.; Chen, L.; Bodner, G. Estimation of runoff mitigation by morphologically different cover crop root systems. J. Hydrol. 2016, 538, 667–676. [Google Scholar] [CrossRef]
- Austin, E.E.; Wickings, K.; McDaniel, M.D.; Robertson, G.P.; Grandy, A.S. Cover crop root contributions to soil carbon in a no-till corn bioenergy cropping system. GCB Bioenergy 2017, 9, 1252–1263. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Bodner, G.; Alsalem, M.; Nakhforoosh, A.; Arnold, T.; Leitner, D. RGB and spectral root imaging for plant phenotyping and physiological research: Experimental setup and imaging protocols. JoVE 2017, 126, e56251. [Google Scholar] [CrossRef]
- Kutschera, L.; Lichtenegger, E. Wurzelatlas mitteleuropäischer Ackerunkräuter und Kulturpflanzen; DLG-Verlag: Frankfurt am Main, Germany, 1960. [Google Scholar]
- Kutschera, L.; Lichtenegger, E.; Sobotik, M. Wurzelatlas der Kulturpflanzen gemäßigter Gebiete mit Arten des Feldgemüsebaues; DLG-Verlag: Frankfurt am Main, Germany, 2009. [Google Scholar]
- Bodner, G.; Leitner, D.; Nakhforoosh, A.; Sobotik, M.; Moder, K.; Kaul, H.-P. A statistical approach to root system classification. Front. Plant Sci. 2013, 4, 292. [Google Scholar] [CrossRef]
- Wojciechowski, T.; Gooding, M.J.; Ramsay, L.; Gregory, P.J. The effects of dwarfing genes on seedling root growth of wheat. J. Exp. Bot. 2009, 60, 2565–2573. [Google Scholar] [CrossRef]
- Watt, M.; Moosavi, S.; Cunningham, S.C.; Kirkegaard, J.A.; Rebetzke, G.J.; Richards, R.A. A rapid, controlled-environment seedling root screen for wheat correlates well with rooting depths at vegetative, but not reproductive, stages at two field sites. Ann. Bot. 2013, 112, 447–455. [Google Scholar] [CrossRef]
- Zhao, J.; Bodner, G.; Rewald, B.; Leitner, D.; Nagel, K.A.; Nakhforoosh, A. Root architecture simulation improves the inference from seedling root phenotyping towards mature root systems. J. Exp. Bot. 2017, 68, 965–982. [Google Scholar] [CrossRef]
- Lobet, G.; Pound, M.P.; Diener, J.; Pradal, C.; Draye, X.; Godin, C.; Javaux, M.; Leitner, D.; Meunier, F.; Nacry, P. Root system markup language: Toward a unified root architecture description language. Plant Physiol. 2015, 167, 617–627. [Google Scholar] [CrossRef]
- Mullen, J.L.; Hangarter, R.P. Genetic analysis of the gravitropic set-point angle in lateral roots of Arabidopsis. Adv. Space Res. 2003, 31, 2229–2236. [Google Scholar] [CrossRef]
- Tian, H.; De Smet, I.; Ding, Z. Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci. 2014, 19, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Cassab, G. Other Tropisms and Their Relationship to Gravitropism. In Plant Tropism, 1st ed.; Gilroy, S., Masson, H., Eds.; Blackwell Publishing: Carlton, Australia, 2008; pp. 123–139. [Google Scholar]
- Pound, M.P.; French, A.P.; Atkinson, J.A.; Wells, D.M.; Bennett, M.J.; Pridmore, T. RootNav: Navigating images of complex root architectures. Plant Physiol. 2013, 162, 1802–1814. [Google Scholar] [CrossRef] [Green Version]
- Belachew, K.Y.; Nagel, K.A.; Fiorani, F.; Stoddard, F.L. Diversity in root growth responses to moisture deficit in young faba bean (Vicia faba L.) plants. PeerJ 2018, 6, e4401. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Forde, B.G. Quantitative analysis of lateral root development: Pitfalls and how to avoid them. Plant Cell 2012, 24, 4–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Pagès, L. Branching patterns of root systems: Quantitative analysis of the diversity among dicotyledonous species. Ann. Bot. 2014, 114, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [Green Version]
- Eissenstat, D.M. Costs and benefits of constructing roots of small diameter. J. Plant Nutr. 1992, 15, 763–782. [Google Scholar] [CrossRef]
- Ostonen, I.; Püttsepp, Ü.; Biel, C.; Alberton, O.; Bakker, M.R.; Lõhmus, K.; Majdi, H.; Metcalfe, D.; Olsthoorn, A.F.M.; Pronk, A. Specific root length as an indicator of environmental change. Plant Biosyst. 2007, 141, 426–442. [Google Scholar] [CrossRef]
- Zobel, R.W. Tertiary Roots Systems. In Roots and Soil Management: Interactions between Roots and the Soil; Zobel, R.W., Wright, S.F., Eds.; Agronomy Monograph 48; American Society of Agronomy-Crop Science Society of America-Soil Science Society of America: Madison, WI, USA, 2005; pp. 35–56. [Google Scholar]
- Schnepf, A.; Leitner, D.; Landl, M.; Lobet, G.; Mai, T.H.; Morandage, S.; Sheng, C.; Zörner, M.; Vanderborght, J.; Vereecken, H. CRootBox: A structural—Functional modelling framework for root systems. Ann. Bot. 2018, 121, 1033–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, H.; Yan, X.; Rubio, G.; Beebe, S.E.; Blair, M.W.; Lynch, J.P. Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Funct. Plant Biol. 2004, 31, 959–970. [Google Scholar] [CrossRef]
- Lyu, Y.; Tang, H.; Li, H.; Zhang, F.; Rengel, Z.; Whalley, W.R.; Shen, J. Major crop species show differential balance between root morphological and physiological responses to variable phosphorus supply. Front. Plant Sci. 2016, 7, 1939. [Google Scholar] [CrossRef] [PubMed]
- Bodner, G.; Leitner, D.; Kaul, H.-P. Coarse and fine root plants affect pore size distributions differently. Plant Soil 2014, 380, 133–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesuffleur, F.; Paynel, F.; Bataillé, M.-P.; Le Deunff, E.; Cliquet, J.-B. Root amino acid exudation: Measurement of high efflux rates of glycine and serine from six different plant species. Plant Soil 2007, 294, 235–246. [Google Scholar] [CrossRef]
- Okubo, A.; Matsusaka, M.; Sugiyama, S. Impacts of root symbiotic associations on interspecific variation in sugar exudation rates and rhizosphere microbial communities: A comparison among four plant families. Plant Soil 2016, 399, 345–356. [Google Scholar] [CrossRef]
- Redin, M.; Guénon, R.; Recous, S.; Schmatz, R.; de Freitas, L.L.; Aita, C.; Giacomini, S.J. Carbon mineralization in soil of roots from twenty crop species, as affected by their chemical composition and botanical family. Plant Soil 2014, 378, 205–214. [Google Scholar] [CrossRef]
- Roumet, C.; Urcelay, C.; Díaz, S. Suites of root traits differ between annual and perennial species growing in the field. New Phytol. 2006, 170, 357–368. [Google Scholar] [CrossRef]
- Rosner, K.; Bodner, G.; Hage-Ahmed, K.; Steinkellner, S. Long-term Soil Tillage and Cover Cropping Affected Arbuscular Mycorrhizal Fungi, Nutrient Concentrations, and Yield in Sunflower. Agron. J. 2018, 110, 2664–2672. [Google Scholar] [CrossRef]
- Thingstrup, I.; Rubaek, G.; Sibbesen, E.; Jakobsen, I. Flax (Linum usitatissimum L.) depends on arbuscular mycorrhizal fungi for growth and P uptake at intermediate but not high soil P levels in the field. Plant Soil 1998, 203, 37–46. [Google Scholar] [CrossRef]
- Brundrett, M.C. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002, 154, 275–304. [Google Scholar] [CrossRef]
- Liu, A.; Ma, B.L.; Bomke, A.A. Effects of cover crops on soil aggregate stability, total organic carbon, and polysaccharides. Soil Sci. Soc. Am. J. 2005, 69, 2041–2048. [Google Scholar] [CrossRef]
- Kabir, Z.; Koide, R.T. The effect of dandelion or a cover crop on mycorrhiza inoculum potential, soil aggregation and yield of maize. Agric. Ecosyst. Environ. 2000, 78, 167–174. [Google Scholar] [CrossRef]
- Lal, R. Soil carbon sequestration and aggregation by cover cropping. J. Soil Water Conserv. 2015, 70, 329–339. [Google Scholar] [CrossRef]
- Breland, T.A. Green manuring with clover and ryegrass catch crops undersown in spring wheat: Effects on soil structure. Soil Use Manag. 1995, 11, 163–167. [Google Scholar] [CrossRef]
- Brown, L.K.; George, T.S.; Neugebauer, K.; White, P.J. The rhizosheath—A potential trait for future agricultural sustainability occurs in orders throughout the angiosperms. Plant Soil 2017, 418, 115–128. [Google Scholar] [CrossRef]
- Teboh, J.M.; Franzen, D.W. Buckwheat (Fagopyrum esculentum Moench) potential to contribute solubilized soil phosphorus to subsequent crops. Commun. Soil. Sci. Plant Anal. 2011, 42, 1544–1550. [Google Scholar] [CrossRef]
- Zhu, Y.G.; He, Y.Q.; Smith, S.E.; Smith, F.A. Buckwheat (Fagopyrum esculentum Moench) has high capacity to take up phosphorus (P) from a calcium (Ca)-bound source. Plant Soil 2002, 239, 1–8. [Google Scholar] [CrossRef]
- Eichler-Löbermann, B.; Gaj, R.; Schnug, E. Improvement of soil phosphorus availability by green fertilization with catch crops. Commun. Soil. Sci. Plant Anal. 2009, 40, 70–81. [Google Scholar] [CrossRef]
- Leitner, D.; Felderer, B.; Vontobel, P.; Schnepf, A. Recovering root system traits using image analysis exemplified by two-dimensional neutron radiography images of lupine. Plant Physiol. 2014, 164, 24–35. [Google Scholar] [CrossRef] [Green Version]


| Species | Total Length (cm) | Top–Bottom Ratio † (relative) | Diameter (mm) | Fine Root Ratio †† (relative) |
|---|---|---|---|---|
| Vicia faba | 570.5 f | 0.051 c | 1.02 b | 0.278 d |
| Vicia sativa | 2075.3 bcd | 0.070 bc | 0.94 bc | 0.336 d |
| Trifolium alexandrinum | 743.4 ef | 0.076 bc | 0.73 cde | 0.498 bc |
| Camelina sativa | 2694.9 b | 0.138 bc | 1.02 b | 0.315 d |
| Raphanus sativus | 6164.8 a | 0.265 a | 0.88 bcd | 0.481 c |
| Fagopyrum esculentum | 1498.1 cde | 0.089 bc | 0.68 de | 0.610 ab |
| Phacelia tanacetifolia | 2245.4 bc | 0.126 bc | 1.01 b | 0.513 bc |
| Helianthus annuus | 2661.7 b | 0.151 b | 1.33 a | 0.331 d |
| Avena strigosa | 1274.8 def | 0.138 bc | 1.02 b | 0.251 d |
| Linum usitatissimum | 1008.0 ef | 0.075 bc | 0.58 e | 0.673 a |
| p-value | <0.001 | 0.005 | <0.001 | <0.001 |
| Species | Lateral Distance (cm) | Lateral Angle † (degree) | Primary-Lateral Ratio †† (rel.) | Convex Hull (cm2) |
|---|---|---|---|---|
| V. faba | 1.3 a | 71.7 bc | 1.67 a | 102.0 abc |
| V. sativa | 1.3 a | 65.5 cd | 1.91 a | 76.3 bc |
| T. alexandrinum | 0.6 b | 59.1 d | 0.28 b | 138.9 abc |
| C. sativa | 0.2 de | 75.3 b | 0.31 b | 128.9 abc |
| R. sativus | 0.2 e | 75.5 b | 0.11 b | 199.3 a |
| F. esculentum | 0.3 cde | 71.4 bc | 0.16 b | 173.5 ab |
| P. tanacetifolia | 0.3 cd | 76.4 b | 0.20 b | 202.9 a |
| H. annuus | 0.4 bc | 70.5 bc | 0.43 b | 199.8 a |
| A. strigosa | 0.6 b | 84.8 a | 2.60 a | 53.1 c |
| L. usitatissimum | 1.3 a | 85.3 a | 2.81 a | 57.7 c |
| p-value | <0.001 | <0.001 | <0.001 | 0.039 |
| Species | Length (cm) | Depth (cm) | Highest Order (Number) |
|---|---|---|---|
| V. faba | 2053.2 | 90 | 4 |
| V. sativa | 1933.1 | 80 | 3 |
| T. alexandrinum | - | - | - |
| C. sativa | 841.1 | 93 | 4 |
| R. sativus | 2946.6 | 153 | 4 |
| F. esculentum | 1730.7 | 105 | 5 |
| P. tanacetifolia | 2061.6 | 88 | 4 |
| H. annuus | 5291.0 | 180 | 4 |
| A. strigosa * | 1265.6 | 92 | 3 |
| L. usitatissimum | 1259.6 | 60 | 3 |
| Species | Family |
|---|---|
| Vicia faba L. (Faba bean) | Leguminoseae |
| Vicia sativa L. (Common vetch) | Leguminoseae |
| Trifolium alexandrinum L. (Berseem clover) 1 | Leguminoseae |
| Camelina sativa (L.) Crantz (Cemelina) | Brassicaceae |
| Raphanus raphanistrum subsp. sativus (L.) Domin (Radish) | Brassicaceae |
| Fagopyrum esculentum Moench (Buckwheat) | Polygonaceae |
| Phacelia tanacetifolia Benth. (Phacelia) | Boraginaceae |
| Helianthus annuus L. (Sunflower) | Asteraceae |
| Avena strigosa Schreb. (Black oat) 1 | Poaceae |
| Linum usitatissimum L. (Linseed) | Linaceae |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodner, G.; Loiskandl, W.; Hartl, W.; Erhart, E.; Sobotik, M. Characterization of Cover Crop Rooting Types from Integration of Rhizobox Imaging and Root Atlas Information. Plants 2019, 8, 514. https://doi.org/10.3390/plants8110514
Bodner G, Loiskandl W, Hartl W, Erhart E, Sobotik M. Characterization of Cover Crop Rooting Types from Integration of Rhizobox Imaging and Root Atlas Information. Plants. 2019; 8(11):514. https://doi.org/10.3390/plants8110514
Chicago/Turabian StyleBodner, Gernot, Willibald Loiskandl, Wilfried Hartl, Eva Erhart, and Monika Sobotik. 2019. "Characterization of Cover Crop Rooting Types from Integration of Rhizobox Imaging and Root Atlas Information" Plants 8, no. 11: 514. https://doi.org/10.3390/plants8110514
APA StyleBodner, G., Loiskandl, W., Hartl, W., Erhart, E., & Sobotik, M. (2019). Characterization of Cover Crop Rooting Types from Integration of Rhizobox Imaging and Root Atlas Information. Plants, 8(11), 514. https://doi.org/10.3390/plants8110514






