New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria That Different?
Abstract
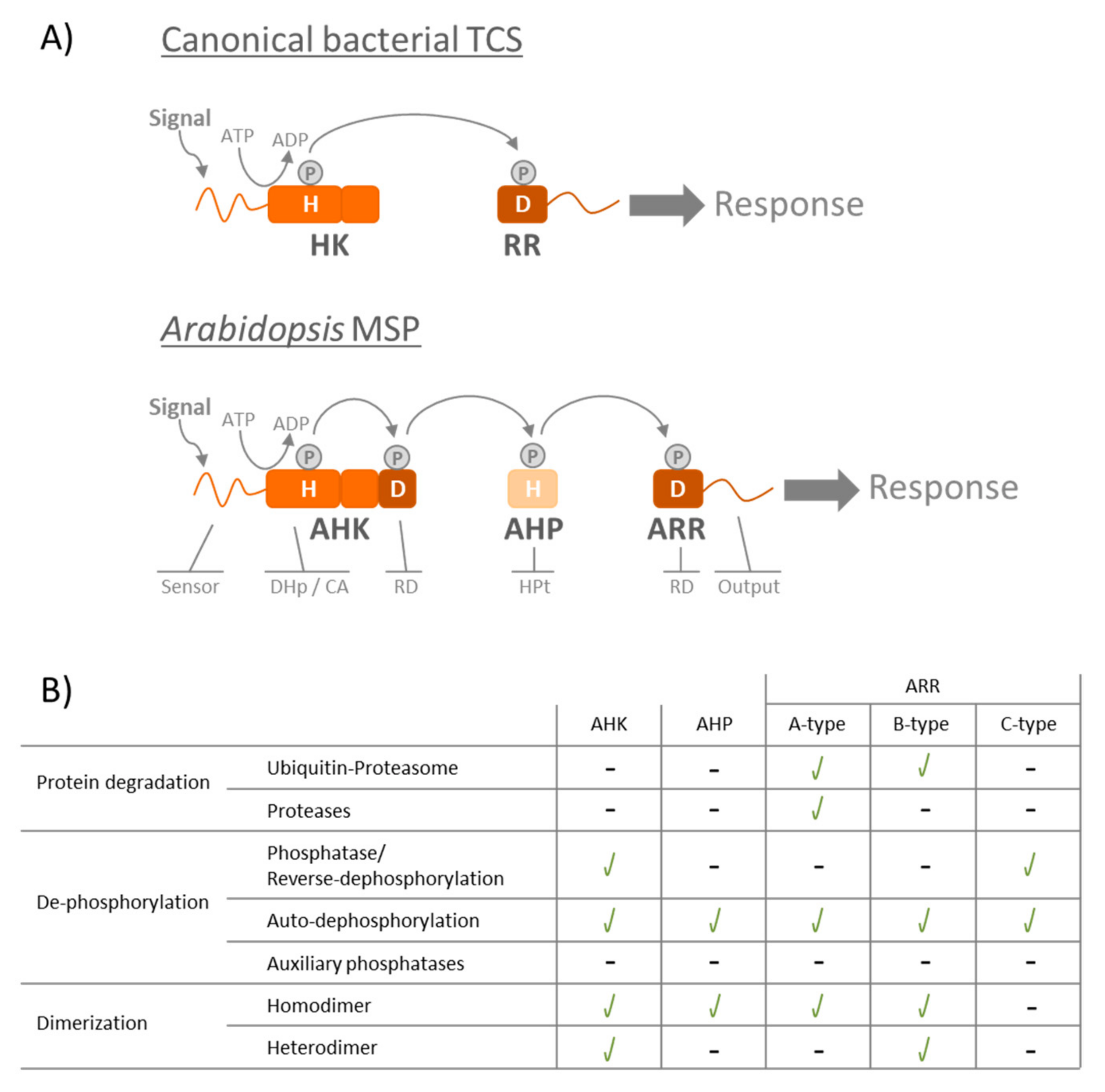
:1. Review
2. TCS/MSP Regulation via Response Regulators’ Protein Degradation
3. TCS/MSP Regulation via Protein Dephosphorylation
4. TCS/MSP Regulation via Protein Dimerization
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TCS | two-component system |
| MSP | multistep-phosphorelay |
| HK | histidine kinase |
| HPt | phosphotransfer protein |
| RR | response regulator |
| RD | receiver domain |
| CA | catalytic domain |
| AHK | Arabidopsis histidine kinase |
| AHP | Arabidopsis His-containing phosphotransfer protein |
| ARR | Arabidopsis response regulators |
| His; H | histidine |
| Asp; D | aspartate |
| DHp | dimerization and histidine phosphotransfer domain |
| HTH | helix-turn-helix |
| SCF | Skp, Cullin, and F-box containing complex |
| KMD | kiss me deadly |
| DEG9 | degradation of periplasmic proteins 9 |
References
- Nixon, B.T.; Ronson, C.W.; Ausubel, F.M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 1986, 8320, 7850–7854. [Google Scholar] [CrossRef] [Green Version]
- Stock, J.B.; Ninfa, A.J.; Stock, A.M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 1989, 534, 450–490. [Google Scholar] [PubMed]
- Bourret, R.B. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010, 132, 142–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galperin, M.Y. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 2006, 18812, 4169–4182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Stock, A.M. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 2010, 132, 160–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Stock, A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef] [Green Version]
- Muller, B. Generic signal-specific responses: Cytokinin and context-dependent cellular responses. J. Exp. Bot. 2011, 6210, 3273–3288. [Google Scholar] [CrossRef]
- Williams, R.H.; Whitworth, D.E. The genetic organisation of prokaryotic two-component system signalling pathways. BMC Genom. 2010, 11, 720. [Google Scholar] [CrossRef] [Green Version]
- Whitworth, D.E. Classification and Organization of Two-component Systems. Two-Component Systems; Horizon Scientific Press: CA, USA, 2012; pp. 1–20. [Google Scholar]
- Casino, P.; Lopez-Redondo, M.; Marina, A. Structural Basis of Signal Transduction and Specificity in Two-component Systems. Two-Component Systems in Bacteria; Caister Academic Press: Wymondham, UK, 2012; pp. 21–40. [Google Scholar]
- Gross, R.; Beier, D. Two-Component Systems in Bacteria; Caister Academic Press: Norfolk, UK, 2012. [Google Scholar]
- Koretke, K.K.; Lupas, A.N.; Warren, P.V.; Rosenberg, M.; Brown, J.R. Evolution of two-component signal transduction. Mol. Biol. Evol. 2000, 1712, 1956–1970. [Google Scholar] [CrossRef] [Green Version]
- Saito, H. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem. Rev. 2001, 1018, 2497–2509. [Google Scholar] [CrossRef]
- Mizuno, T. Two-component phosphorelay signal transduction systems in plants: From hormone responses to circadian rhythms. Biosci. Biotechnol. Biochem. 2005, 6912, 2263–2276. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi, C.; Koizumi, H.; Suzuki, T.; Mizuno, T. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001, 422, 231–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, H.; Suzuki, T.; Terada, K.; Takei, K.; Ishikawa, K.; Miwa, K.; Yamashino, T.; Mizuno, T. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001, 429, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi., C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 166, 1365–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, J.; Sakai, H.; Nourizadeh, S.; Chen, Q.G.; Bleecker, A.B.; Ecker, J.R.; Meyerowitz, E.M. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 1998, 108, 1321–1332. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.H.; Yoo, S.D. ETHYLENE RESPONSE 1 histidine kinase activity of Arabidopsis promotes plant growth. Plant Physiol. 2007, 1432, 612–616. [Google Scholar] [CrossRef] [Green Version]
- Grefen, C.; Städele, K.; Růžička, K.; Obrdlik, P.; Harter, K.; Horák, J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 2008, 12, 308–320. [Google Scholar] [CrossRef]
- Kumar, M.N.; Jane, W.N.; Verslues, P. Role of the putative osmosensor Arabidopsis Histidine Kinase 1 (AHK1) in dehydration avoidance and low water potential response. Plant Physiol. 2013, 16, 942–953. [Google Scholar] [CrossRef] [Green Version]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 2008, 204, 1101–1117. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.S.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought.; salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 10451, 20623–20628. [Google Scholar] [CrossRef] [Green Version]
- Pischke, M.S.; Jones, L.G.; Otsuga, D.; Fernandez, D.E.; Drews, G.N.; Sussman, M.R. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 2002, 9924, 15800–15805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O.; Ku, S.-J.; Cho, C.; Lee, D.J.; Lee, E.-J.; Strnad, M.; et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 28530, 23371–23386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 246, 2578–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vescovi, M.; Riefler, M.; Gessuti, M.; Novák, O.; Schmülling, T.; Schiavo, F.L. Programmed cell death induced by high levels of cytokinin in Arabidopsis cultured cells is mediated by the cytokinin receptor CRE1/AHK4. J. Exp. Bot. 2012, 637, 2825–2832. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.; Desikan, R. Modulation of ROS production and hormone levels by AHK5 during abiotic and biotic stress signaling. Plant Signal. Behav. 2012, 78, 893–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desikan, R.; Horák, J.; Chaban, C.; Mira-Rodado, V.; Witthöft, J.*; Elgass, K.; Grefen, C.; Cheung, M.-K.; Meixner, A.J.; Hooley, R.; et al. The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS ONE 2008, 36, e2491. [Google Scholar] [CrossRef] [Green Version]
- Mira-Rodado, V.; Veerabagu, M.; Witthöft, J.; Teply, J.; Harter, K.; Desikan, R. Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Signal. Behav. 2012, 711, 1467–1476. [Google Scholar] [CrossRef] [Green Version]
- Eisele, J.F.; Fäßler, F.; Bürgel, P.F.; Chaban, C. A Rapid and Simple Method for Microscopy-Based Stomata Analyses. PLoS ONE 2016, 1110, e0164576. [Google Scholar] [CrossRef]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000, 246, 703–711. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef]
- Imamura, A.; Kiba, T.; Tajima, Y.; Yamashino, T.; Mizuno, T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003, 442, 122–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 1711, 3007–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veerabagu, M.; Elgass, K.; Kirchler, T.; Huppenberger, P.; Harter, K.; Chaban, C.; Mira-Rodado, V. The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J. 2012, 725, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Veerabagu, M.; Kirchler, T.; Elgass, K.; Stadelhofer, B.; Stahl, M.; Harter, K.; Mira-Rodado, V.; Chaban, C. The Interaction of the Arabidopsis Response Regulator ARR18 with bZIP63 Mediates the Regulation of PROLINE DEHYDROGENASE Expression. Mol. Plant 2014, 710, 1560–1577. [Google Scholar] [CrossRef] [Green Version]
- Wallmeroth, N.; Anastasia, A.K.; Harter, K.; Berendzen, K.W.; Mira-Rodado, V. Arabidopsis response regulator 22 inhibits cytokinin-regulated gene transcription in vivo. Protoplasma 2017, 2541, 597–601. [Google Scholar] [CrossRef]
- Wallmeroth, N.; Jeschke, D.; Slane, D.; Nägele, J.; Veerabagu, M.; Mira-Rodado, V.; Berendzen, K.W. ARR22 overexpression can suppress plant Two-Component Regulatory Systems. PLoS ONE 2019, 142, e0212056. [Google Scholar] [CrossRef]
- Haydon, M.J.; Mielczarek, O.; Robertson, F.C.; Hubbard, K.E.; Webb, A.A.R. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 2013, 502, 689–692. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 1102, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Yamashino, T.; Mizuno, T. Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 7211, 3025–3029. [Google Scholar] [CrossRef] [Green Version]
- Salomé, P.A.; To, J.P.; Kieber, J.J.; McClung, C.R. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 2006, 181, 55–69. [Google Scholar]
- Lohar, D.P.; Schaff, J.E.; Laskey, J.G.; Kieber, J.J.; Bilyeu, K.D.; Bird, D.M. Cytokinins play opposite roles in lateral root formation.; nematode and Rhizobial symbioses. Plant J. 2004, 382, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Nitschke, S.; Klaumünzer, M.; AbdelGawad, H.; Asard, H.; Grimm, B.; Riefler, M.; Schmülling, T. A novel protective function for cytokinin in the light stress response is mediated by the Arabidopsis histidine kinase2 and Arabidopsis histidine kinase3 receptors. Plant Physiol. 2014, 1643, 1470–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.; Kim, J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3.; AHP5 function in cold signaling. Plant Physiol. 2013, 1611, 408–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, N.Y.; Cho, C.; Kim, J. Inducible expression of Arabidopsis response regulator 22 (ARR22), a type-C ARR, in transgenic Arabidopsis enhances drought and freezing tolerance. PLoS ONE 2013, 811, e79248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; To, J.P.C.; Cheng, C.-Y.; Schaller, G.E.; Kieber, J.J. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 2011, 681, 1–10. [Google Scholar] [CrossRef]
- Hass, C.; Lohrmann, J.; Albrecht, V.; Sweere, U.; Hummel, F.; Yoo, S.D.; Hwang, I.; Zhu, T.; Schäfer, E.; Kudla, J.; et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 2004, 2316, 3290–3302. [Google Scholar] [CrossRef] [Green Version]
- Kushwah, S.; Laxmi, A. The interaction between glucose and cytokinin signal transduction pathway in Arabidopsis thaliana. Plant Cell Environ. 2014, 371, 235–253. [Google Scholar] [CrossRef]
- Mira-Rodado, V.; Sweere, U.; Grefen, C.; Kunkel, T.; Fejes, E.; Nagy, F.; Schäfer, E.; Harter, K. Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J. Exp. Bot. 2007, 5810, 2595–2607. [Google Scholar] [CrossRef]
- Sweere, U.; Eichenberg, K.; Lohrmann, J.; Mira-Rodado, V.; Bäurle, I.; Kudla, J.; Nagy, F.; Schäfer, E.; Harter, K. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 2001, 294, 1108–1111. [Google Scholar] [CrossRef] [Green Version]
- Buechel, S.; Leibfried, A.; To, J.P.; Zhao, Z.; Andersen, S.U.; Kieber, J.J.; Lohmann, J.U. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 2010, 89, 279–284. [Google Scholar] [CrossRef]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.-C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 224, 1232–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gattolin, S.; Alandete-Saez, M.; Elliott, K.; Gonzalez-Carranza, Z.; Naomab, E.; Powell, C.; Roberts, J.A. Spatial and temporal expression of the response regulators ARR22 and ARR24 in Arabidopsis thaliana. J. Exp. Bot. 2006, 5715, 4225–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horák, J.; Grefen, C.; Berendzen, K.W.; Hahn, A.; Stierhof, Y.-D.; Stadelhofer, B.; Stahl, M.; Koncz, C.; Harter, K. The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 2008, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiba, T.; Aoki, K.; Sakakibara, H.; Mizuno, T. Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 2004, 458, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Sheen, J. Advances in cytokinin signaling. Science 2007, 318, 68–69. [Google Scholar] [CrossRef] [PubMed]
- Caesar, K.; Thamm, A.M.K.; Witthöft, J.; Elgass, K.; Huppenberger, P.; Grefen, C.; Horak, J.; Harter, K. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J. Exp. Bot. 2011, 6215, 5571–5580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wulfetange, K.; Lomin, S.N.; Romanov, G.A.; Stolz, A.; Heyl, A.; Schmülling, T. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011, 1564, 1808–1818. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.J.; Kim, S.; Ha, Y.-M.; Kim, J. Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta 2008, 2273, 577–587. [Google Scholar] [CrossRef]
- Kiba, T.; Yamada, H.; Sato, S.; Kato, T.; Tabata, S.; Yamashino, T.; Mizuno, T. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003, 448, 868–874. [Google Scholar] [CrossRef] [Green Version]
- D’Agostino, I.B.; Deruere, J.; Kieber, J.J. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 1244, 1706–1717. [Google Scholar]
- To, J.P.; Deruère, J.; Maxwell, B.B.; Morris, V.F.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 2007, 1912, 3901–3914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchison, C.E.; Kieber, J.J. Signaling via Histidine-Containing Phosphotransfer Proteins in Arabidopsis. Plant Signal. Behav. 2007, 24, 287–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seshasayee, A.S.; Luscombe, N.M. Comparative genomics suggests differential deployment of linear and branched signaling across bacteria. Mol. Biosyst. 2011, 711, 3042–3049. [Google Scholar] [CrossRef]
- Laub, M.T.; Goulian, M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007, 41, 121–145. [Google Scholar] [CrossRef] [Green Version]
- Kunst, F.; Msadek, T.; Bignon, J.; Rapoport, G. The DegS/DegU and ComP/ComA two-component systems are part of a network controlling degradative enzyme synthesis and competence in Bacillus subtilis. Res. Microbiol. 1994, 145, 393–402. [Google Scholar] [CrossRef]
- Mader, U.; Antelmann, H.; Buder, T.; Dahl, M.; Hecker, M.; Homuth, G. Bacillus subtilis functional genomics: Genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genom. 2002, 2684, 455–467. [Google Scholar] [CrossRef]
- Ogura, M.; Yamaguchi, H.; Yoshida, K.; Fujita, Y.; Tanaka, T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: An approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Res. 2001, 2918, 3804–3813. [Google Scholar] [CrossRef] [Green Version]
- Ogura, M.; Tsukahara, K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol. Microbiol. 2010, 755, 1244–1259. [Google Scholar] [CrossRef]
- Iniesta, A.A.; Shapiro, L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. USA 2008, 10543, 16602–16607. [Google Scholar] [CrossRef] [Green Version]
- Jenal, U.; Fuchs, T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998, 1719, 5658–5669. [Google Scholar] [CrossRef] [Green Version]
- Collier, J. Regulation of chromosomal replication in Caulobacter crescentus. Plasmid 2012, 672, 76–87. [Google Scholar] [CrossRef]
- Biondi, E.G.; Reisinger, S.J.; Skerker, J.M.; Arif, M.; Perchuk, B.S.; Ryan, K.R.; Laub, M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 2006, 444, 899–904. [Google Scholar] [CrossRef]
- Angelastro, P.S.; Sliusarenko, O.; Jacobs-Wagner, C. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J. Bacteriol. 2010, 1922, 539–552. [Google Scholar] [CrossRef] [Green Version]
- Quon, K.C.; Yang, B.; Domian, I.J.; Shapiro, L.; Marczynski, G.T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 1998, 951, 120–125. [Google Scholar] [CrossRef] [Green Version]
- Domian, I.J.; Quon, K.C.; Shapiro, L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 1997, 903, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, C.; Ausmees, N.; Cordwell, S.J.; Shapiro, L.; Laub, M.T. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 2003, 475, 1279–1290. [Google Scholar] [CrossRef]
- Iniesta, A.A.; McGrath, P.T.; Reisenauer, A.; McAdams, H.H.; Shapiro, L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA 2006, 10329, 10935–10940. [Google Scholar] [CrossRef] [Green Version]
- Butler, S.M.; Festa, R.A.; Pearce, M.J.; Darwin, K.H. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 2006, 603, 553–562. [Google Scholar] [CrossRef]
- Sauer, R.T.; Baker, T.A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011, 80, 587–612. [Google Scholar] [CrossRef]
- Frees, R.; Chastanet, A.; Qazi, S.; Sørensen, K.; Hill, P.; Msadek, T.; Ingmer, H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 2004, 545, 1445–1462. [Google Scholar] [CrossRef]
- Maurizi, M.R.; Thompson, M.W.; Singh, S.K.; Kim, S.-H. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzym. 1994, 244, 314–331. [Google Scholar]
- Goldberg, A.L.; Moerschell, R.P.; Hachung, C.; Maurizi, M.R. ATP-dependent protease La (lon) from Escherichia coli. Methods Enzym. 1994, 244, 350–375. [Google Scholar]
- Darwin, K.H. Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nat. Rev. Microbiol. 2009, 77, 485–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Cinos, C.; Goossens, K.; Salado, I.G.; Van Der Veken, P.; De Winter, H.; Augustyns, K. ClpP Protease, a Promising Antimicrobial Target. Int. J. Mol. Sci. 2019, 20, 2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, R.S.; Vierstra, R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 1961, 13–28. [Google Scholar] [CrossRef]
- Kim, K.; Ryu, H.; Cho, Y.-H.; Scacchi, E.; Sabatini, S.; Hwang, I. Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR 2 attenuates signaling output in two-component circuitry. Plant J. 2012, 696, 934–945. [Google Scholar] [CrossRef]
- Kim, K.; Hwang, I. Attenuation of cytokinin signaling via proteolysis of a type-B response regulator. Plant Signal. Behav. 2012, 77, 756–759. [Google Scholar] [CrossRef] [Green Version]
- Kurepa, J.; Li, Y.; Smalle, J.A. Cytokinin signaling stabilizes the response activator ARR1. Plant J. 2014, 781, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Shull, T.E.; Kurepa, J.; Smalle, J.A. Cytokinin signaling promotes differential stability of type-B ARRs. Plant Signal. Behav. 2016, 114, e1169354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Chiang, Y.-H.; Kieber, J.J.; Schaller, G.E. SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. USA 2013, 11024, 10028–10033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitsma, J.M.; Liu, X.; Reichermeier, K.M.; Moradian, A.; Sweredoski, M.J.; Hess, S.; Deshaies, R.J. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell 2017, 1716, 1326–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Kurepa, J.; Smalle, J. AXR1 promotes the Arabidopsis cytokinin response by facilitating ARR5 proteolysis. Plant J. 2013, 741, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Li, Y.; Smalle, J.A. Proteasome-dependent proteolysis has a critical role in fine-tuning the feedback inhibition of cytokinin signaling. Plant Signal. Behav. 2013, 83, e23474. [Google Scholar]
- Smalle, J.; Kurepa, J.; Yang, P.; Babiychuk, E.; Kushnir, S.; Durski, A.; Vierstra, R.D. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 2002, 141, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Smalle, J.; Kurepa, J.; Yang, P.; Emborg, T.J.; Babiychuk, E.; Kushnir, S.; Vierstra, R.D. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 2003, 154, 965–980. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 106, 385–397. [Google Scholar] [CrossRef]
- Van der Hoorn, R.A. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [Green Version]
- Chi, W.; Li, J.; He, B.; Chai, X.; Xu, X.; Sun, X.; Jiang, J.; Feng, P.; Zuo, J.; Lin, R.; et al. DEG9, a serine protease, modulates cytokinin and light signaling by regulating the level of ARABIDOPSIS RESPONSE REGULATOR 4. Proc. Natl. Acad. Sci. USA 2016, 11325, E3568–E3576. [Google Scholar] [CrossRef] [Green Version]
- Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 2011, 123, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. How important is the phosphatase activity of sensor kinases? Curr. Opin. Microbiol. 2010, 132, 168–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, R.C. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 1997, 368, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, P.M.; Webre, D.J.; Stock, J.B. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry 2003, 4247, 14075–14082. [Google Scholar] [CrossRef] [PubMed]
- Perego, M.; Hanstein, C.; Welsh, K.M.; Djavakhishvili, T.; Glaser, P.; Hoch, J.A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 1994, 796, 1047–1055. [Google Scholar] [CrossRef]
- Silversmith, R.E. Auxiliary phosphatases in two-component signal transduction. Curr. Opin. Microbiol. 2010, 132, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Igo, M.M.; Ninfa, A.J.; Stock, J.B.; Silhavy, T.J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989, 311, 1725–1734. [Google Scholar] [CrossRef] [Green Version]
- Igo, M.M.; Ninfa, A.J.; Silhavy, T.J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989, 35, 598–605. [Google Scholar] [CrossRef] [Green Version]
- Jin, T.; Inouye, M. Transmembrane signaling. Mutational analysis of the cytoplasmic linker region of Taz1-1, a Tar-EnvZ chimeric receptor in Escherichia coli. J. Mol. Biol. 1994, 2445, 477–481. [Google Scholar] [CrossRef]
- Slauch, J.M.; Silhavy, T.J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 1989, 2102, 281–292. [Google Scholar] [CrossRef]
- Groban, E.S.; Clarke, E.J.; Salis, H.M.; Miller, S.M.; Voigt, C.A. Kinetic buffering of cross talk between bacterial two-component sensors. J. Mol. Biol. 2009, 3903, 380–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Qin, L.; Yoshida, T.; Inouye, M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl. Acad. Sci. USA 2000, 9714, 7808–7813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.J.; Inouye, M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002, 27727, 24155–24161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, S.T.; Kenney, L.J. Application of fluorescence resonance energy transfer to examine EnvZ/OmpR interactions. Methods Enzym. 2007, 422, 352–360. [Google Scholar]
- Weiss, V.; Magasanik, B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 8523, 8919–8923. [Google Scholar] [CrossRef] [Green Version]
- Skarphol, K.; Waukau, J.; Forst, S.A. Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. J. Bacteriol. 1997, 1794, 1413–1416. [Google Scholar] [CrossRef] [Green Version]
- Jagadeesan, S.; Mann, P.; Schink, C.W.; Higgs, P.I. A novel “four-component” two-component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J. Biol. Chem. 2009, 28432, 21435–21445. [Google Scholar] [CrossRef] [Green Version]
- Kamberov, E.S.; Atkinson, M.R.; Chandran, P.; Ninfa, A.J. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J. Biol. Chem. 1994, 26945, 28294–28299. [Google Scholar]
- Sheeler, N.L.; MacMillan, S.V.; Nodwell, J.R. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J. Bacteriol. 2005, 1872, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Georgellis, D.; Kwon, O.; De Wulf, P.; Lin, E.C.C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J. Biol. Chem. 1998, 27349, 32864–32869. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Liu, W.; Hulett, F.M. Decay of activated Bacillus subtilis pho response regulator, PhoP approximately P, involves the PhoR approximately P intermediate. Biochemistry 1999, 3831, 10119–10125. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.D.; Wolanin, P.M.; Stock, J.B. Signal transduction in bacterial chemotaxis. Bioessays 2006, 281, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.; Oosawa, K.; Kaplan, N.; Simon, M. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 1988, 531, 79–87. [Google Scholar] [CrossRef]
- Wylie, D.; Stock, A.; Wong, C.-Y.; Stock, J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem. Biophys. Res. Commun. 1988, 1512, 891–896. [Google Scholar] [CrossRef]
- Zapf, J.; Madhusudan; Grimshaw, C.E.; Hoch, J.A.; Varughese, K.I.; Whiteley, J.M. A source of response regulator autophosphatase activity: The critical role of a residue adjacent to the Spo0F autophosphorylation active site. Biochemistry 1998, 3721, 7725–7732. [Google Scholar] [CrossRef] [PubMed]
- Lukat, G.S.; Stock, A.M.; Stock, J.B. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry 1990, 2923, 5436–5442. [Google Scholar] [CrossRef] [PubMed]
- Appleby, J.L.; Bourret, R.B. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 1998, 18014, 3563–3569. [Google Scholar]
- Lukat, G.S.; Lee, B.H.; Mottonen, J.M.; Stock, A.M.; Stock, J.B. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 1991, 26613, 8348–8354. [Google Scholar]
- Ninfa, A.J. Phosphatase Activity of Two-component System Transmitter Proteins. In Two-Component Systems in Bacteria; Caister Academic Press: Wymondham, UK, 2012; pp. 85–108. [Google Scholar]
- Silversmith, R.E.; Levin, M.D.; Schilling, E.; Bourret, R.B. Kinetic characterization of catalysis by the chemotaxis phosphatase CheZ. Modulation of activity by the phosphorylated CheY substrate. J. Biol. Chem. 2008, 2832, 756–765. [Google Scholar] [CrossRef] [Green Version]
- Mähönen, A.P.; Higuchi, M.; Törmäkangas, K.; Miyawaki, K.; Pischke, M.S.; Sussman, M.R.; Helariutta, Y.; Kakimoto, T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 2006, 1611, 1116–1122. [Google Scholar]
- Nakamura, A.; Kakimoto, T.; Imamura, A.; Suzuki, T.; Ueguchi, C.; Mizuno, T. Biochemical characterization of a putative cytokinin-responsive His-kinase, CKI1, from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 1999, 639, 1627–1630. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Imamura, A.; Ueguchi, C.; Mizuno, T. Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol. 1998, 3912, 1258–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Kiba, T.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999, 407, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Tatsuno, I.; Lee, L.; Kawagishi, I.; Homma, M.; Imae, Y. Transmembrane signalling by the chimeric chemosensory receptors of Escherichia coli Tsr and Tar with heterologous membrane-spanning regions. Mol. Microbiol. 1994, 144, 755–762. [Google Scholar] [CrossRef]
- Kofoid, E.C.; Parkinson, J.S. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. USA 1988, 8514, 4981–4985. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Saha, S.K.; Tomomori, C.; Ishima, R.; Liu, D.; Tong, K.I.; Park, H.; Dutta, R.; Qin, L.; Swindells, M.B.; et al. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature 1998, 396, 88–92. [Google Scholar] [CrossRef]
- Tomomori, C.; Tanaka, T.; Dutta, R.; Park, H.; Saha, S.K.; Zhu, Y.; Ishima, R.; Liu, D.; I Tong, K.; Kurokawa, H.; et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct Biol. 1999, 68, 729–734. [Google Scholar]
- Marina, A.; Waldburger, C.D.; Hendrickson, W.A. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005, 2424, 4247–4259. [Google Scholar] [CrossRef]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3128, ra50. [Google Scholar] [CrossRef] [Green Version]
- Bhate, M.P.; Molnar, K.S.; Goulian, M.; DeGrado, W.F. Signal transduction in histidine kinases: Insights from new structures. Structure 2015, 236, 981–994. [Google Scholar] [CrossRef] [Green Version]
- Ashenberg, O.; Keating, A.E.; Laub, M.T. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J. Mol. Biol. 2013, 4257, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Swanson, R.V.; Bourret, R.B.; Simon, M.I. Intermolecular complementation of the kinase activity of CheA. Mol. Microbiol. 1993, 83, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Casino, P.; Rubio, V.; Marina, A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 2009, 1392, 325–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena-Sandoval, G.R.; Georgellis, D. The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction. J. Bacteriol. 2010, 1926, 1735–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dortay, H.; Mehnert, N.; Bürkle, L.; Schmülling, T.; Heyl, A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006, 27320, 4631–4644. [Google Scholar] [CrossRef]
- Schaller, G.E.; Ladd, A.N.; Lanahan, M.B.; Spanbauer, J.M.; Bleecker, A.B. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 1995, 27021, 12526–12530. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Wen, C.-K.; Binder, B.M.; Chen, Y.-F.; Chang, J.; Chiang, Y.-H.; Kerris, R.J.; Chang, C.; Schaller, G.E. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J. Biol. Chem. 2008, 28335, 23801–23810. [Google Scholar] [CrossRef] [Green Version]
- Obrdlik, P.; El-Bakkoury, M.; Hamacher, T.; Cappellaro, C.; Vilarino, C.; Fleischer, C.; Ellerbrok, H.; Kamuzinzi, R.; Ledent, V.; Blaudez, D.; et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 2004, 10133, 12242–12247. [Google Scholar] [CrossRef] [Green Version]
- Mayerhofer, H.; Panneerselvam, S.; Kaljunen, H.; Tuukkanen, A.; Mertens, H.D.T.; Mueller-Dieckmann, J. Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1). J. Biol. Chem. 2015, 2905, 2644–2658. [Google Scholar] [CrossRef] [Green Version]
- Pekárová, B.; Szmitkowska, A.; Dopitová, R.; Degtjarik, O.; Žídek, L.; Hejátko, J. Structural Aspects of Multistep Phosphorelay-Mediated Signaling in Plants. Mol. Plant 2016, 91, 71–85. [Google Scholar]
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 132, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Volz, K. Structural conservation in the CheY superfamily. Biochemistry 1993, 3244, 11741–11753. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Cho, H.S.; Pelton, J.G.; Yan, D.; Berry, E.A.; Wemmer, D.E. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 2001, 27619, 16425–16431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y.; Torre, A.D.L.; Yan, D.; Kustu, S.; Nixon, B.T.; Wemmer, D.E. Regulation of the transcriptional activator NtrC1: Structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003, 1720, 2552–2563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachhawat, P.; Swapna, G.; Montelione, G.T.; Stock, A.M. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 2005, 139, 1353–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiedler, U.; Weiss, V. A common switch in activation of the response regulators NtrC and PhoB: Phosphorylation induces dimerization of the receiver modules. EMBO J. 1995, 1415, 3696–3705. [Google Scholar] [CrossRef]
- McCleary, W.R. The activation of PhoB by acetylphosphate. Mol. Microbiol. 1996, 206, 1155–1163. [Google Scholar] [CrossRef]
- Jeon, Y.; Lee, Y.S.; Han, J.S.; Kim, J.B.; Hwang, D.S. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 2001, 27644, 40873–40879. [Google Scholar] [CrossRef] [Green Version]
- Robinson, V.L.; Wu, T.; Stock, A.M. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J. Bacteriol. 2003, 18514, 4186–4194. [Google Scholar] [CrossRef] [Green Version]
- Toro-Roman, A.; Mack, T.R.; Stock, A.M. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: A symmetric dimer mediated by the alpha4-beta5-alpha5 face. J. Mol. Biol. 2005, 3491, 11–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro-Roman, A.; Wu, T.; Stock, A.M. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005, 1412, 3077–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassmann, P.; Chan, C.; Paul, R.; Beck, A.; Heerklotz, H.; Jenal, U.; Schirmer, T. Structure of BeF3− -modified response regulator PleD: Implications for diguanylate cyclase activation, catalysis.; feedback inhibition. Structure 2007, 158, 915–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mack, T.R.; Gao, R.; Stock, A.M. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J. Mol. Biol. 2009, 3892, 349–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, K.; Shinagawa, H.; Amemura, M.; Kawamoto, T.; Yamada, M.; Nakata, A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 1989, 2103, 551–559. [Google Scholar] [CrossRef]
- Shinagawa, H.; Makino, K.; Nakata, A. Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J. Mol. Biol. 1983, 1683, 477–488. [Google Scholar] [CrossRef]
- Sola, M.; Gomis-Rüth, F.X.; Serrano, L.; González, A.; Coll1, M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 1999, 2852, 675–687. [Google Scholar] [CrossRef]
- Wilson, D.; Pethica, R.; Zhou, Y.; Talbot, C.; Vogel, C.; Madera, M.; Chothia, C.; Gough, J. SUPERFAMILY-sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009, 37, D380–D386. [Google Scholar] [CrossRef]
- Muller-Dieckmann, H.J.; Grantz, A.A.; Kim, S.H. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure 1999, 712, 1547–1556. [Google Scholar] [CrossRef] [Green Version]
- Pekárová, B.; Klumpler, T.; Třísková, O.; Jansen, S.; Dopitová, R.; Borkovcová, P.; Papoušková, V.; Hejátko, J.; Horák, J.; Nejedlá, E.; et al. Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana. Plant J. 2011, 675, 827–839. [Google Scholar]
- Bauer, J.; Reiss, K.; Veerabagu, M.; Heunemann, M.; Harter, K.; Stehle, T. Structure-function analysis of Arabidopsis thaliana histidine kinase AHK5 bound to its cognate phosphotransfer protein AHP1. Mol. Plant 2013, 63, 959–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosoda, K.; Imamura, A.; Katoh, E.; Hatta, T.; Tachiki, M.; Yamada, H.; Mizuno, T.; Yamazaki, T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 2002, 149, 2015–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Mack, T.R.; Stock, A.M. Bacterial response regulators: Versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007, 325, 225–234. [Google Scholar] [CrossRef] [Green Version]
- Blanco, A.G.; Solà, M.; Gomis-Rüth, F.; Coll, M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002, 105, 701–713. [Google Scholar] [CrossRef]
- Pristovšek, P.; Sengupta, K.; Lohr, F.; Schäfer, B.; Von Trebra, M.W.; Rüterjans, H.; Bernhard, F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 2003, 27820, 17752–17759. [Google Scholar]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 2018, 117, 970–982. [Google Scholar] [CrossRef] [Green Version]
- Scharein, B.; Groth, G. Phosphorylation alters the interaction of the Arabidopsis phosphotransfer protein AHP1 with its sensor kinase ETR1. PLoS ONE 2011, 69, e24173. [Google Scholar] [CrossRef]
- Punwani, J.A.; Hutchison, C.E.; Schaller, G.E.; Kieber, J.J. The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. 2010, 623, 473–482. [Google Scholar] [CrossRef]
- Schaller, G.E.; Shiu, S.H.; Armitage, J.P. Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 2011, 219, R320–R330. [Google Scholar] [CrossRef] [Green Version]
- Horák, J.; Janda, L.; Pekárová, B.; Hejátko, J. Molecular mechanisms of signalling specificity via phosphorelay pathways in Arabidopsis. Curr. Protein Pept. Sci. 2011, 122, 126–136. [Google Scholar]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mira-Rodado, V. New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria That Different? Plants 2019, 8, 590. https://doi.org/10.3390/plants8120590
Mira-Rodado V. New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria That Different? Plants. 2019; 8(12):590. https://doi.org/10.3390/plants8120590
Chicago/Turabian StyleMira-Rodado, Virtudes. 2019. "New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria That Different?" Plants 8, no. 12: 590. https://doi.org/10.3390/plants8120590
APA StyleMira-Rodado, V. (2019). New Insights into Multistep-Phosphorelay (MSP)/Two-Component System (TCS) Regulation: Are Plants and Bacteria That Different? Plants, 8(12), 590. https://doi.org/10.3390/plants8120590



