The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele
Abstract
:1. Introduction
2. Results
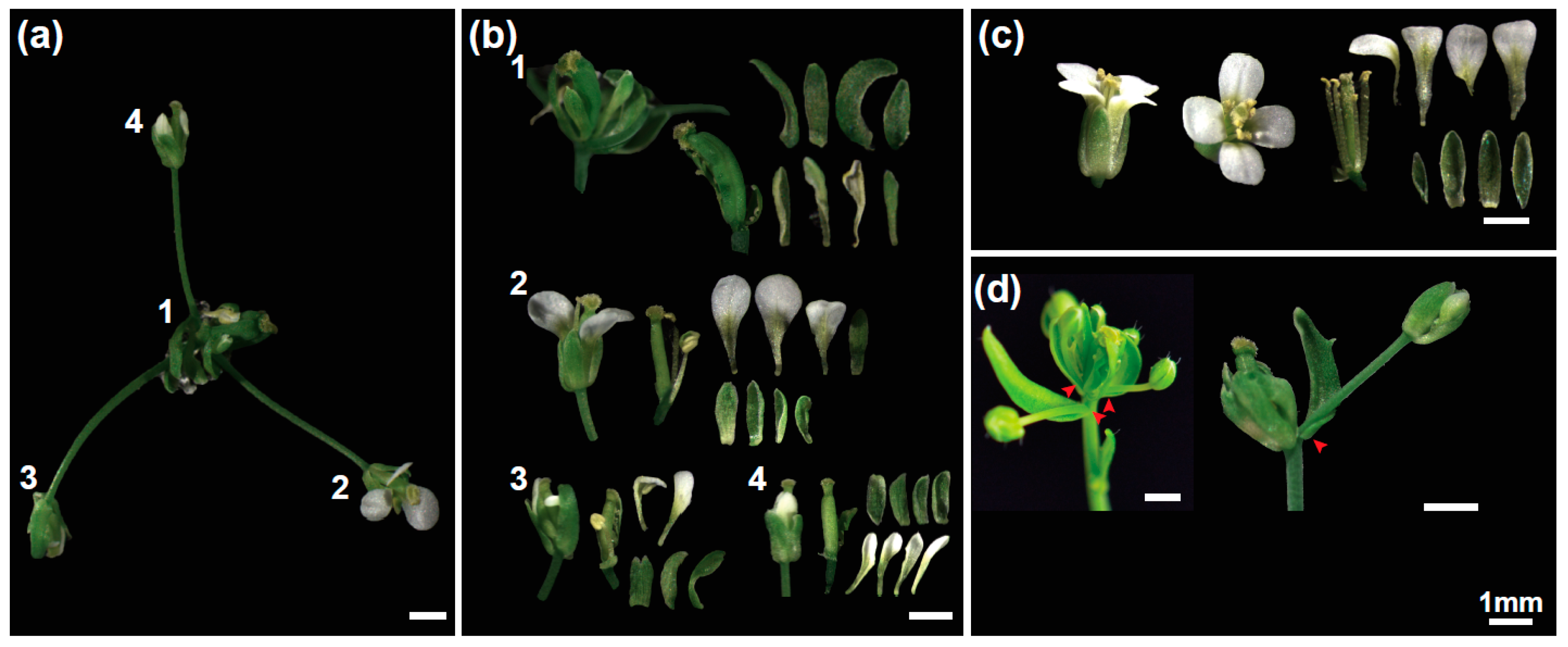
2.1. The Flower-in-Flower (fif) Transposon Insertion Line Displays a Novel Flower Phenotype
2.2. The Transposon Insertion is Not Responsible for the fif Phenotype
2.3. The fif Phenotype is Caused by a Novel Allele of LEAFY (LFY)
2.4. LFYFIF Impairs DNA-Binding Capability but Shows Wild Type Intracellular Localization and Homomerization
3. Discussion
4. Materials and Methods
4.1. Plant Material and Arabidopsis thaliana Transformation
4.2. Plasmid Construction
4.3. Classical Mapping and Mapping by Genome Sequencing
4.4. Localization and FRET-FLIM Studies
4.5. qDPI-ELISA, DPI-ELISA Based Screening and Western Blotting
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pineiro, M.; Coupland, G. The control of flowering time and floral identity in Arabidopsis. Plant Physiol. 1998, 117, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H. To be, or not to be, a flower–control of floral meristem identity. Trends Genet. 1998, 14, 26–32. [Google Scholar] [CrossRef]
- Pidkowich, M.S.; Klenz, J.E.; Haughn, G.W. The making of a flower: Control of floral meristem identity in Arabidopsis. Trends Plant Sci. 1999, 4, 64–70. [Google Scholar] [CrossRef]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef] [PubMed]
- O’Maoileidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prunet, N. My favourite flowering image: An Arabidopsis inflorescence expressing fluorescent reporters for the APETALA3 and SUPERMAN genes. J. Exp. Bot. 2018. [Google Scholar] [CrossRef]
- Wagner, D. Key developmental transitions during flower morphogenesis and their regulation. Curr. Opin. Genet. Dev. 2017, 45, 44–50. [Google Scholar] [CrossRef]
- Wellmer, F.; Graciet, E.; Riechmann, J.L. Specification of floral organs in Arabidopsis. J. Exp. Bot. 2014, 65, 1–9. [Google Scholar] [CrossRef]
- Irish, V.F.; Sussex, I.M. Function of the Apetala-1 Gene during Arabidopsis Floral Development. Plant Cell 1990, 2, 741–753. [Google Scholar] [CrossRef] [Green Version]
- Huala, E.; Sussex, I.M. Leafy Interacts with Floral Homeotic Genes to Regulate Arabidopsis Floral Development. Plant Cell 1992, 4, 901–913. [Google Scholar] [CrossRef]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E.M.; Smyth, D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef] [Green Version]
- Mandel, M.A.; Gustafsonbrown, C.; Savidge, B.; Yanofsky, M.F. Molecular Characterization of the Arabidopsis Floral Homeotic Gene Apetala1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Kempin, S.A.; Savidge, B.; Yanofsky, M.F. Molecular-Basis of the Cauliflower Phenotype in Arabidopsis. Science 1995, 267, 522–525. [Google Scholar] [CrossRef]
- Blazquez, M.A. Illuminating flowers: CONSTANS induces LEAFY expression. Bioessays 1997, 19, 277–279. [Google Scholar] [CrossRef]
- Moyroud, E.; Minguet, E.G.; Ott, F.; Yant, L.; Pose, D.; Monniaux, M.; Blanchet, S.; Bastien, O.; Thevenon, E.; Weigel, D.; et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 2011, 23, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Winter, C.M.; Austin, R.S.; Blanvillain-Baufume, S.; Reback, M.A.; Monniaux, M.; Wu, M.F.; Sang, Y.; Yamaguchi, A.; Yamaguchi, N.; Parker, J.E.; et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 2011, 20, 430–443. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Weigel, D. Building beauty: The genetic control of floral patterning. Dev. Cell 2002, 2, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Shinya, T.; Galis, I.; Narisawa, T.; Sasaki, M.; Fukuda, H.; Matsuoka, H.; Saito, M.; Matsuoka, K. Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol. 2007, 48, 1404–1413. [Google Scholar] [CrossRef] [Green Version]
- Miwa, H.; Betsuyaku, S.; Iwamoto, K.; Kinoshita, A.; Fukuda, H.; Sawa, S. The Receptor-Like Kinase SOL2 Mediates CLE Signaling in Arabidopsis. Plant Cell Physiol. 2008, 49, 1752–1757. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Wada, T.; Kamiya, A.; Yuguchi, M.; Yokouchi, T.; Imura, Y.; Takabe, H.; Sakurai, T.; Akiyama, K.; Hirayama, T.; et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006, 47, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Motohashi, R.; Kuromori, T.; Mizukado, S.; Sakurai, T.; Kanahara, H.; Seki, M.; Shinozaki, K. A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002, 129, 1695–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Hirayama, T.; Kiyosue, Y.; Takabe, H.; Mizukado, S.; Sakurai, T.; Akiyama, K.; Kamiya, A.; Ito, T.; Shinozaki, K. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004, 37, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Kover, P.X.; Valdar, W.; Trakalo, J.; Scarcelli, N.; Ehrenreich, I.M.; Purugganan, M.D.; Durrant, C.; Mott, R. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000551. [Google Scholar] [CrossRef] [Green Version]
- Pacurar, D.I.; Pacurar, M.L.; Street, N.; Bussell, J.D.; Pop, T.I.; Gutierrez, L.; Bellini, C. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 2012, 63, 2491–2501. [Google Scholar] [CrossRef] [Green Version]
- James, G.V.; Patel, V.; Nordstrom, K.J.V.; Klasen, J.R.; Salome, P.A.; Weigel, D.; Schneeberger, K. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013, 14, R61. [Google Scholar] [CrossRef] [Green Version]
- Schneeberger, K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014, 15, 662–676. [Google Scholar] [CrossRef]
- Wu, X.; Dinneny, J.R.; Crawford, K.M.; Rhee, Y.; Citovsky, V.; Zambryski, P.C.; Weigel, D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 2003, 130, 3735–3745. [Google Scholar] [CrossRef] [Green Version]
- Siriwardana, N.S.; Lamb, R.S. A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana. Plant J. 2012, 71, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.M.; Böser, A.; Hirsch, J.P.; Wanke, D. Quantitative Analysis of Protein-DNA Interaction by qDPI-ELISA. Methods Mol. Biol. 2016, 1482, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.H.; Henneges, C.; Schussler, A.; Kolukisaoglu, H.U.; Koch, G.; Wallmeroth, N.; Hecker, A.; Thurow, K.; Zell, A.; Harter, K.; et al. Screening for protein-DNA interactions by automatable DNA-protein interaction ELISA. PLoS ONE 2013, 8, e75177. [Google Scholar] [CrossRef] [Green Version]
- Weigel, D.; Nilsson, O. A developmental switch sufficient for flower initiation in diverse plants. Nature 1995, 377, 495–500. [Google Scholar] [CrossRef]
- Sayou, C.; Nanao, M.H.; Jamin, M.; Pose, D.; Thevenon, E.; Gregoire, L.; Tichtinsky, G.; Denay, G.; Ott, F.; Peirats Llobet, M.; et al. A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat. Commun. 2016, 7, 11222. [Google Scholar] [CrossRef] [Green Version]
- William, D.A.; Su, Y.; Smith, M.R.; Lu, M.; Baldwin, D.A.; Wagner, D. Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 2004, 101, 1775–1780. [Google Scholar] [CrossRef] [Green Version]
- Hames, C.; Ptchelkine, D.; Grimm, C.; Thevenon, E.; Moyroud, E.; Gerard, F.; Martiel, J.L.; Benlloch, R.; Parcy, F.; Muller, C.W. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 2008, 27, 2628–2637. [Google Scholar] [CrossRef] [Green Version]
- Chahtane, H.; Vachon, G.; Le Masson, M.; Thevenon, E.; Perigon, S.; Mihajlovic, N.; Kalinina, A.; Michard, R.; Moyroud, E.; Monniaux, M.; et al. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013, 74, 678–689. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Clark, R.M.; Lanz, C.; Warthmann, N.; Weigel, D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008, 18, 2024–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneeberger, K.; Hagmann, J.; Ossowski, S.; Warthmann, N.; Gesing, S.; Kohlbacher, O.; Weigel, D. Simultaneous alignment of short reads against multiple genomes. Genome Biol. 2009, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.Q.; Schneeberger, K. SHOREmap v3.0: Fast and Accurate Identification of Causal Mutations from Forward Genetic Screens. In Plant Functional Genomics: Methods and Protocols, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1284, pp. 381–395. [Google Scholar]
- Schneeberger, K.; Ossowski, S.; Lanz, C.; Juul, T.; Petersen, A.H.; Nielsen, K.L.; Jorgensen, J.E.; Weigel, D.; Andersen, S.U. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 2009, 6, 550–551. [Google Scholar] [CrossRef]
- Ladwig, F.; Dahlke, R.I.; Stuhrwohldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine Regulates Growth in Arabidopsis through a Response Module at the Plasma Membrane That Includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef] [Green Version]
- Ohmi, Y.; Ise, W.; Harazono, A.; Takakura, D.; Fukuyama, H.; Baba, Y.; Narazaki, M.; Shoda, H.; Takahashi, N.; Ohkawa, Y.; et al. Sialylation converts arthritogenic IgG into inhibitors of collagen-induced arthritis. Nat. Commun. 2016, 7, 11205. [Google Scholar] [CrossRef] [Green Version]
- Brand, L.H.; Kirchler, T.; Hummel, S.; Chaban, C.; Wanke, D. DPI-ELISA: A fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 2010, 6, 25. [Google Scholar] [CrossRef] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohrholz, A.; Sun, H.; Glöckner, N.; Hummel, S.; Kolukisaoglu, Ü.; Schneeberger, K.; Harter, K. The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele. Plants 2019, 8, 599. https://doi.org/10.3390/plants8120599
Mohrholz A, Sun H, Glöckner N, Hummel S, Kolukisaoglu Ü, Schneeberger K, Harter K. The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele. Plants. 2019; 8(12):599. https://doi.org/10.3390/plants8120599
Chicago/Turabian StyleMohrholz, Anne, Hequan Sun, Nina Glöckner, Sabine Hummel, Üner Kolukisaoglu, Korbinian Schneeberger, and Klaus Harter. 2019. "The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele" Plants 8, no. 12: 599. https://doi.org/10.3390/plants8120599
APA StyleMohrholz, A., Sun, H., Glöckner, N., Hummel, S., Kolukisaoglu, Ü., Schneeberger, K., & Harter, K. (2019). The Striking Flower-in-Flower Phenotype of Arabidopsis thaliana Nossen (No-0) is Caused by a Novel LEAFY Allele. Plants, 8(12), 599. https://doi.org/10.3390/plants8120599





