RETRACTED: Morpho-Physiological and Proteomic Analyses of Eucalyptus camaldulensis as a Bioremediator in Copper-Polluted Soil in Saudi Arabia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Material Collection and Treatment
2.3. Growth and Physiological Performance
2.4. Determination of Relative Water Content
2.5. Determination of Chlorophyll Content
2.6. Determination of Copper (Cu) Concentration
2.7. Protein Extraction from Plant Leaves
2.8. Trichloroacetic Acid (TCA)/Acetone Protocol
2.9. Two-Dimensional Gel Electrophoresis (2-DE) Protein Gel Electrophoresis
2.10. Protein Imaging
2.11. Bioinformatics Analysis of the Identified Proteins
2.12. Statistical Analysis
3. Results and Discussion
3.1. Changes in Plant Morphology
3.2. Influence of Cu on Plant Growth
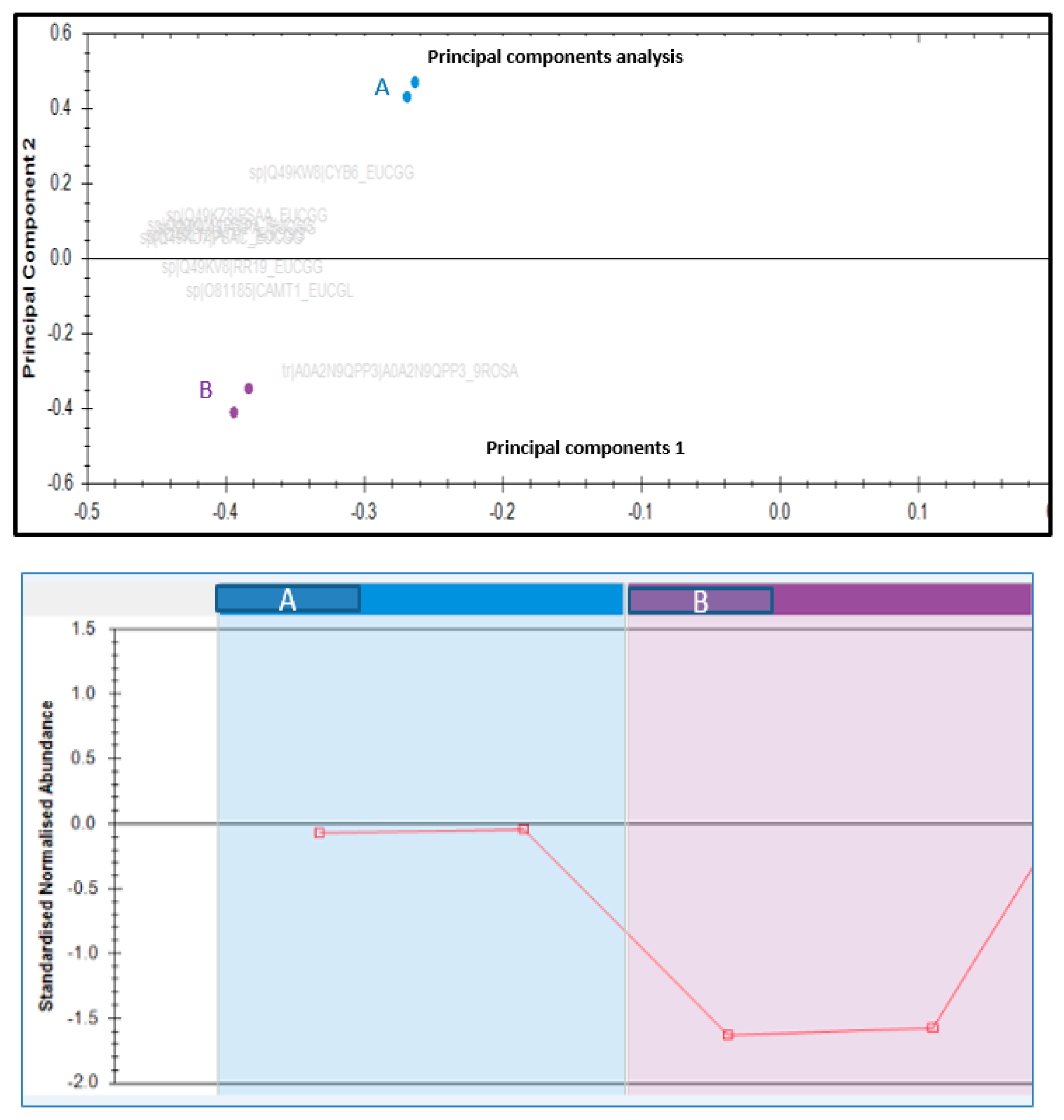
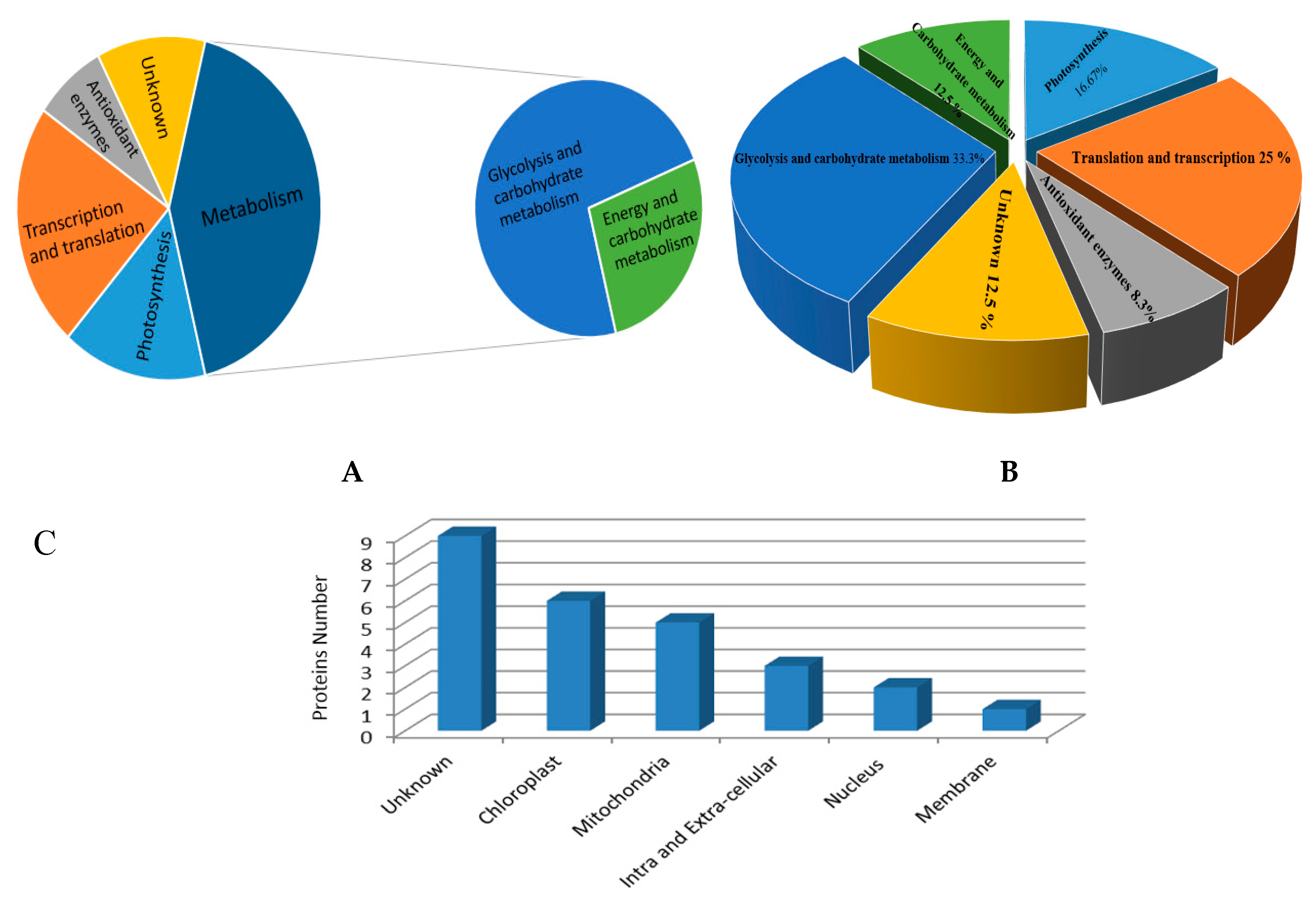
3.3. Classification of Identified Proteins
3.4. Changes in Proteomic Profile
3.4.1. Photosynthetic Proteins
3.4.2. Antioxidant Enzymes and Related Proteins
3.4.3. Glycolysis and Carbohydrate Metabolism Related Proteins
3.4.4. Energy and Carbohydrate Metabolism Proteins
3.4.5. Proteins Involved in Transcription and Translation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sathiyamoorthy, P.; Damme, P.V.; Oven, M.; Golan-Goldhirsh, A. Heavy metals in medicinal and fodder plants of the Negev desert. J. Environ. Sci. Health 1997, 32, 2111–2123. [Google Scholar] [CrossRef]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M. Hyper accumulators, arbuscular mycorrhizal fungi and stress of heavy metal. Biotechnol. Adv. 2011, 29, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, J.S. Heavy Metals. J. Chem. Edu. 1997, 74, 1374. [Google Scholar] [CrossRef]
- Lenntech. Water Treatment and Air Purification Water Treatment; Lenntech: Rotterdamseweg, The Netherlands, 2004. [Google Scholar]
- Chehregani, A.; Malayeri, B.; Golmohammadi, R. Effects of heavy metals on the developmental stages of ovules and embryonic sac in Euphorbia cheirandenia. Pak. J. Biol. Sci. 2005, 8, 622–625. [Google Scholar]
- Rai, P.K. Impacts of particulate matter pollution on plants: Implications for environmental biomonitoring. Ecotoxicol. Environ. Saf. 2016, 129, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Arena, C.; Maio, A.D.; Nicola, F.D.; Santorufo, L.; Vitale, L.; Maisto, G. Assessment of Eco-Physiological Performance of Quercus ilex L. Leaves in Urban Area by an Integrated Approach. Water Air Soil Pollut. 2014, 225, 1824. [Google Scholar] [CrossRef]
- Maisto, G.; Santorufo, L.; Arena, C. Heavy metal accumulation in leaves affects physiological performance and litter quality of Quercus ilex L. J. Plant Nutr. Soil Sci. 2013, 176, 776–784. [Google Scholar] [CrossRef]
- Jjemba, P.K. Environmental Microbiology: Principles and Applications; Science Publishers Inc.: Enfield, NH, USA, 2004. [Google Scholar]
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; 889p. [Google Scholar]
- Ducic, T.; Polle, A. Transport and detoxification of manganese and copper in plants. Braz. J. Plant Physiol. 2005, 17, 103–112. [Google Scholar] [CrossRef]
- Yruela, I. Copper in plants. Braz. J. Plant Physiol. 2005, 17, 145–156. [Google Scholar] [CrossRef]
- Elobeid, M.; Polle, A. Response of grey poplar (Populus x canescens) to copper stress. Plant Stress 2010, 4, 82–86. [Google Scholar]
- Sommer, A.L. Copper as an essential for plant growth. Plant Physiol. 1931, 6, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Strawn, D.G.; Baker, L.L. Speciation of Cu in a contaminated agricultural soil measured by XAFS, µ-XAFS, and µ-XRF. Environ. Sci. Technol. 2008, 42, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front. Plant Sci. 2015, 6, 1143. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis 1998, 19, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Yu, J. An integrated proteomic and metabolomics study on the chronic effects of mercury in Suaeda salsa under an environmentally relevant salinity. PLoS ONE 2013, 8, e64041. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Renaut, J.; Komatsu, S. Recent developments in the application of proteomics to the analysis of plant responses to heavy metal. Proteomics 2009, 9, 2602–2621. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chengcai, C. Towards understanding plant response to heavy metal stress. In AbioticStress in Plants—Mechanisms and Adaptations; Shanker, A., Ed.; Tech Europe: Rijeka, Croatia, 2011; pp. 59–78. [Google Scholar]
- Sharmin, S.A.; Alam, I.; Kim, K.H.; Kim, Y.G.; Kim, P.J.; Bahk, J.D. Chromium-induced physiological and proteomic alterations in roots of Miscanthus sinensis. Plant Sci. 2012, 187, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Marsano, F.; Cavaletto, M.; Berta, G. Proteomic characterization of copper stress response in Cannabis sativa roots. Plant Proteom. 2007. [Google Scholar] [CrossRef] [PubMed]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Cui, J.; Zhang, H.; Wang, G.; Zhao, F.-J.; Shen, Z. Proteomic analysis of copper stress responses in the roots of two rice (Oryza sativa L.) varieties differing in Cu tolerance. Plant Soil 2013, 366, 647–658. [Google Scholar] [CrossRef]
- Hego, E.; Vilain, S.; Barré, A.; Claverol, S.; Dupuy, J.W.; Lalanne, C.; Bonneu, M.; Plomion, C.; Mench, M. Copper stress-induced changes in leaf soluble proteome of Cu-sensitive and tolerant Agrostis capillaris L. populations. Proteomics 2016, 16, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Wang, X.; Xia, Y.; Chen, C.; Shen, Z.; Chen, Y. Proteomic analysis on roots of Oenothera glazioviana under copper-stress conditions. Sci. Rep. 2017, 7, 10589. [Google Scholar] [CrossRef] [PubMed]
- Madejoan, P.; Marañoan, T.; Navarro-FernaÂndez, C.M.; DomõÂnguez, M.T.; Alegre, J.M.; Robinson, B.; Murillo, J.M. Potential of Eucalyptus camaldulensis for phyto-stabilization and biomonitoring of trace element contaminated soils. PLoS ONE 2017, 12, e0180240. [Google Scholar] [CrossRef]
- Assareh, M.H.; Shariat, A.; Ghamari-Zare, A. Seedling response of three Eucalyptus species to copper and zinc toxic concentrations. Caspian J. Environ. Sci. 2018, 6, 97–103. [Google Scholar]
- Benyó, D.; Horváth, E.; Németh, E.; Leviczky, T.; Takács, K.; Lehotai, N.; Feigl, G.; Kolbert, Z.; Ördög, A.; Gallé, R.; et al. Physiological and molecular responses to heavy metal stresses suggest diferent detoxifcation mechanism of Populus deltoides and P. x canadensis. J. Plant Physiol. 2016, 201, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.M. Osmoregulation and water stress in higher plants. Ann Rev Plant Physiol 1984, 35, 299–319. [Google Scholar] [CrossRef]
- Hegedûs, A.; Erdei, S.; Horváth, G. Comparative studies of H2O2 detoxifying enzymes in green and greening barley seedlings under cadmium stress. Plant Sci. 2001, 160, 1085–1093. [Google Scholar]
- Fu, L.; Chen, C.; Wang, B.; Zhou, X.; Li, S.; Guo, P.; Shen, Z.; Wang, G.; Chen, Y. Differences in copper absorption and accumulation between copper-exclusion and copper-enrichment plants: A comparison of structure and physiological responses. PLoS ONE 2015, 10, e0133424. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Cho, S.W.; Kwon, S.J.; Kamal, A.H.; Kim, S.W.; Oh, M.W.; Lee, M.S.; Chung, K.Y.; Xin, Z.; Woo, S.H. Morpho-Physiological and Proteome Level Responses to Cadmium Stress in Sorghum. PLoS ONE 2016. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Weng, Z.X.; Wang, L.X.; Tan, F.I.; Huang, L.; Xing, J.H.; Chen, S.P.; Cheng, C.L.; Chen, W. Proteomic and physiological analyses reveal detoxification and antioxidation induced by Cd stress in Kandelia candel roots. Trees 2013, 27, 583–595. [Google Scholar] [CrossRef]
- Nowicka, B.; Pluciński, B.; Kuczyńska, P.; Kruk, J. Physiological characterization of Chlamydomonas reinhardtii acclimated to chronic stress induced by Ag, Cd, Cr, Cu and Hg ions. Ecotoxicol. Environ. Saf. 2016, 130, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, M.; Abdalla, M.; Elobeid, M. Influence of copper stress on the growth performance of Eucalyptus camaldulensis seedlings. Int. J. Anim. Environ. Sci. 2014, 4, 649–653. [Google Scholar]
- Ahsan, N.; Nakamura, T.; Komatsu, S. Differential responses of microsomal proteins and metabolites in two contrasting cadmium (Cd)-accumulating soybean cultivars under Cd stress. Amino Acids 2012, 42, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Bazihizina, N.; Colzi, I.; Giorni, E.; Mancuso, S.; Gonnelli, C. Photosynthesizing on metal excess: Copper differently induced changes in various photosynthetic parameters in copper tolerant and sensitive Silene paradoxa L. populations. Plant Sci. 2015, 232, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mostofa, M.G.; Fujita, M. Salicylic acid alleviates copper toxicity in rice (Oryza sativa L.) seedlings by up-regulating antioxidative and glyoxalase systems. Ecotoxicology 2013, 22, 959–973. [Google Scholar] [CrossRef] [PubMed]
- Thounaojam, T.C.; Panda, P.; Mazumdar, P.; Kumar, D.; Sharma, G.D.; Sahoo, L.; Panda, S.K. Excess copper induced oxidative stress and response of antioxidants in rice. Plant Physiol. Biochem. 2012, 53, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Bouazizi, H.; Jouili, H.; Geitmann, A.; El Ferjani, E. Cell wall accumulation of Cu ions and modulation of lignifying enzymes in primary leaves of bean seedlings exposed to excess copper. Biol. Trace Elem. Res. 2011, 139, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Cho, S.W.; Kwon, S.J.; Kamal, A.H.M.; Lee, D.G.; Sarker, K.; Lee, M.S.; Xin, Z.; Woo, S.H. Proteome characterization of copper stress response in root of sorghum. Biometals 2017, 30, 765–785. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, Y.; Zhuang, K.; Li, L.; Xia, Y.; Shen, Z. Proteomic Analysis of Copper-Binding Proteins in Excess Copper-Stressed Roots of Two Rice (Oryza sativa L.) Varieties with Different Cu Tolerances. PLoS ONE 2015, 10, e0125367. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, Y.; Yang, Y.; Yang, S.; Sun, X.; Yang, Y. Physiological and Proteomics Analyses Reveal the Mechanism of Eichhornia crassipes Tolerance to High-Concentration Cadmium Stress Compared with Pistia stratiotes. PLoS ONE 2015, 10, e0124304. [Google Scholar] [CrossRef] [PubMed]
- Llorens, N.; Arola, L.; Bladé, C.; Mas, A. Effects of copper exposure upon nitrogen metabolism in tissue cultured Vitis vinifera. Plant Sci. 2000, 160, 159–163. [Google Scholar] [CrossRef]
- Burkhead, J.L.; Reynolds, K.A.G.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Taamalli, M.; Gevi, F.; Timperio, A.M.; Zolla, L.; Ghnaya, T. Cadmium Stress Responses in Brassica juncea: Hints from Proteomics and Metabolomics. J. Proteome Res. 2013, 12, 4979–4997. [Google Scholar] [CrossRef] [PubMed]
- Hossain, Z.; Hajika, M.; Komatsu, S. Comparative proteome analysis of high and low cadmium accumulating soybeans under cadmium stress. Amino Acids 2012, 43, 2393–2416. [Google Scholar] [CrossRef] [PubMed]
- Figlioli, F.; Sorrentino, M.C.; Memoli, V.; Arena, C.; Maisto, G.; Giordano, S.; Capozzi, F.; Spagnuolo, V. Overall plant responses to Cd and Pb metal stress in maize: Growth pattern, ultrastructure, and photosynthetic activity. Environ. Sci. Pollut. Res. 2019, 26, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, M.C.; Capozzi, F.; Amitrano, C.; Giordano, S.; Arena, C.; Spagnuolo, V. Performance of three cardoon cultivars in an industrial heavy metal contaminated soil: Effects on morphology, cytology and photosynthesis. J. Hazard. Mater. 2018, 351, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Mattie, M.D.; Freedman, J.H. Copper-inducible transcription: Regulation by metal-and oxidative stressresponsive pathways. Am. J. Physiol. Cell Physiol. 2004, 286, C293–C301. [Google Scholar] [CrossRef] [PubMed]
- Iannelli, M.A.; Pietrini, F.; Fiore, L.; Petrilli, L.; Massacci, A. Antioxidant response to cadmium in Phragmites australis plants. Plant Physiol. Biochem. 2002, 40, 977–982. [Google Scholar] [CrossRef]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F.; Xie, Y. Heavy Metal Stress and Some Mechanisms of Plant Defense Response Hindawi Publishing Corporation. Sci. World J. 2015, 2015, 756120. [Google Scholar] [CrossRef] [PubMed]
- Melanie, H.; Marcel, I.; Brandán, P.; Jörg, B.; Malek, S.; Vu, V.L.; Sandra, M.; Dörte, B.; Leonardo, A.R.; Lorenz, A.; et al. The glyceraldehyde-3-phosphate dehydrogenase GapDH of Corynebacterium diphtheriae is redox-controlled by protein Smycothiolation under oxidative stress. Sci. Rep. 2017, 7, 5020. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Henson, D.; Nyirenda, M.; Desikan, R.; Harrison, J.; Lewis, M.; Hughes, J.; Neill, S.J. Proteomic identification of glyceraldehydes 3-phosphate dehydrogenase as aninhibitory target of hydrogenperoxide in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Roth, U.; von Roepenack-Lahaye, E.; Clemens, S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J. Exp. Bot. 2006, 57, 4003–4013. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Kathiresan, A.; Bennett, J.; Takabe, T. Comparative tran-scriptome analyses of barley and rice under salts tress. Theor. Appl. Genet. 2006, 112, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, P.; Planchon, S.; Oufir, M.; Ziebel, J.; Dommes, J.; Hoffmann, L.; Hausman, J.F.; Renaut, J. Combining Proteomics, and Metabolite Analyses to Unravel Cadmium Stress-Response in Poplar Leaves. J. Proteome Res. 2009, 8, 400–417. [Google Scholar] [CrossRef] [PubMed]
- Setsuko, K.; AbuH, M.K.; Zahed, H. Wheat proteomics: Proteome modulation and abiotic stress acclimation. Front. Plant Sci. 2014. [Google Scholar] [CrossRef]
- Ritter, A.; Ubertini, M.; Romac, S.; Gaillard, F.; Delage, L.; Mann, A.; Cock, J.M.; Tonon, T.; Correa, J.A.; Potin, P. Copper stress proteomics highlights local adaptation of two strains of the model brown alga Ectocarpus siliculosus. Proteomics 2010, 10, 2074–2088. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.H.; Cho, K.; Kim, D.E.; Uozumi, N.; Chung, K.Y.; Lee, S.Y.; Choi, J.S.; Cho, S.W.; Shin, C.S.; Woo, S.H. Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol. Biol. Rep. 2012, 39, 9059–9074. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.W.; Cen, Z.Y.; Yan, X.; Bian, Y.W.; Deng, X.; Yan, Y.M. Integrated physiological and proteomic analysis reveals underlying response and defense mechanisms of Brachypodium distachyon seedling leaves under osmotic stress, cadmium and their combined stresses. J. Proteom. 2018, 170, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorina, O.S. Functioning of malate dehydrogenase system in mesophyll and bundle sheath cells of maize leaves under salt stress conditions. Russ. J. Plant Physiol. 2007, 54, 728–735. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Harper, J.F. Pumping with plant P-type ATPases. J. Exp. Bot. 1999, 50, 883–893. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Salama, K.H.A.; Al-Mutawa, M.M. Transport proteins and salt tolerance in plants. Plant Sci. 2003, 164, 891–900. [Google Scholar] [CrossRef]
- Gould, G.W.; Cuenda, A.; Thomson, F.; Cohen, P. The activation of distinct mitogen-activated protein kinase cascades is required for the stimulation of 2-deoxyglucose uptake by interleukin-1 and insulinlike growth factor-1 in KB cells. Biochem. J. 1995, 311, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Duguay, J.; Ma, F.; Wang, T.W.; Tshin, R.; Hopkins, M.T.; McNamara, L.; Thompson, J. Modulation of eIF5A1 expression alters xylem abundance in Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Parkash, J.; Vaidya, T.; Kirti, S.; Dutt, S. Translation initiation factor 5A in Picrorhiza is up-regulated during leaf senescence and in response to abscisic acid. Gene 2014, 542, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice Ribosomal Protein Large Subunit Genes and Their Spatio-temporal and Stress Regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, K.; Kieffer, P.; Dommes, J.; Hausman, J.-F.; Renaut, J. Proteomic changes in leaves of poplar exposed to both cadmium and low-temperature. Environ. Exp. Bot. 2014, 106, 112–123. [Google Scholar] [CrossRef]
- Feng, L.; Jiyan, S.; Chaofeng, S.; Guangcun, C.; Shaoping, H.; Yingxu, C. Proteomic characterization of copper stress response in Elsholtzia splendens roots and leaves. Plant Mol. Biol. 2009, 71, 251–263. [Google Scholar] [CrossRef]
- Ahlquist, P. Review RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 2002, 296, 1270–1273. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.; Yao, X.; Yin, J.; Zhang, D.; Ma, H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 2009, 447, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM, pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef] [PubMed]
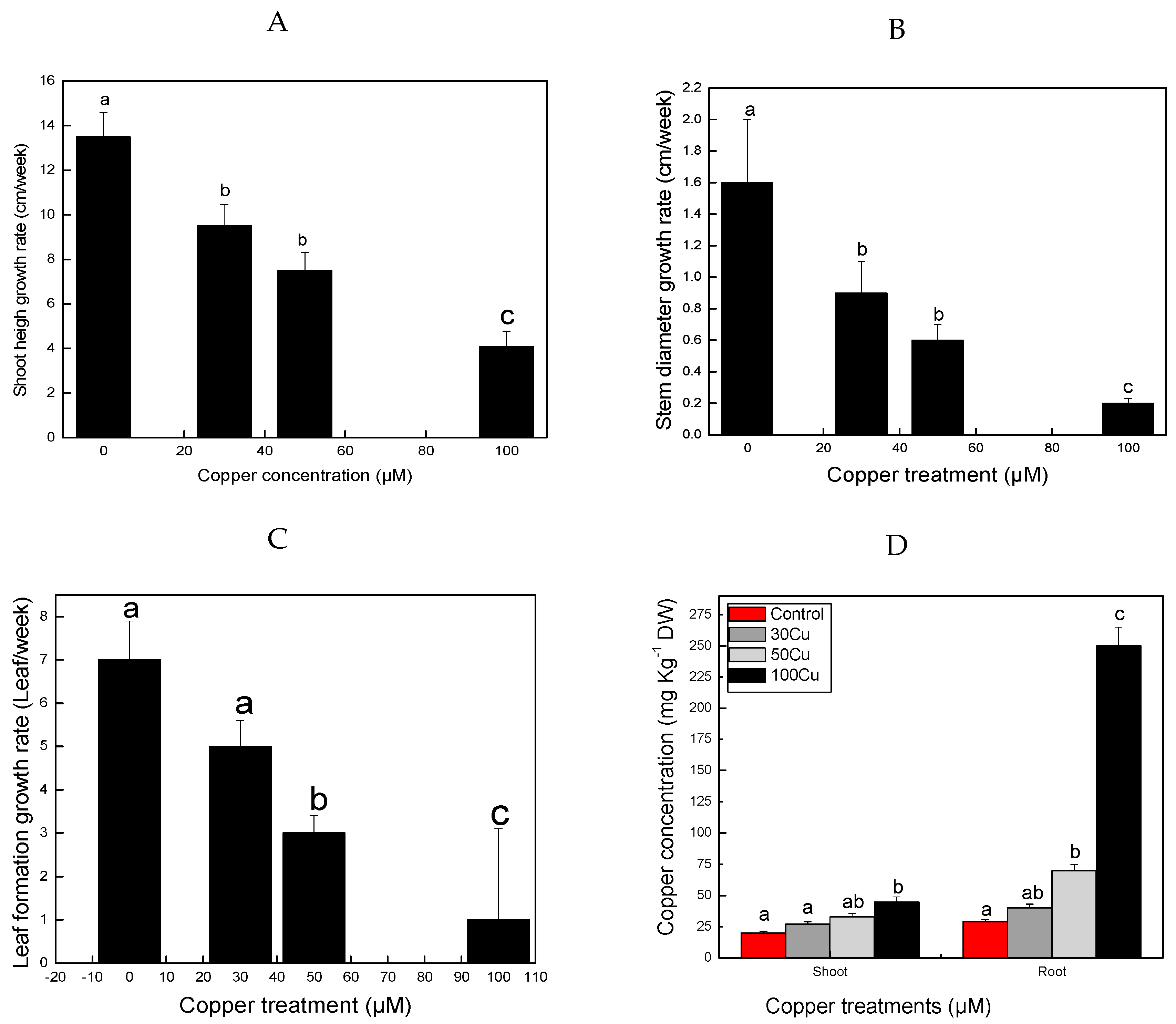

| Physiological Index | Control | Cu (30 µM) | Cu (50 µM) | Cu (100 µM) | Change Fold (Control/Cu 100 µM) |
|---|---|---|---|---|---|
| Relative water content (%) | 84 ± 2.5 | 73 ± 3.1 | 70 ± 2.9 | 56 ± 2.6 * | 1.5 |
| Chlorophyll content (mg/g) | 3.1 ± 0.12 | 2.5 ± 0.3 | 2.1 ± 0.1 | 1.4 ± 0.1 * | 2.21 |
| Shoot fresh weight (g·plant−1) | 2.00 ± 0.01 | 1.98 ± 0.1 | 1.65 ± 0.01 | 1.27 ± 0.09 * | 1.57 |
| Root fresh weight (g·plant−1) | 1.01 ± 0.01 | 0.92 ± 0.01 | 0.72 ± 0.02 | 0.58 ± 0.01 * | 1.74 |
| Shoot dry weight (g·plant−1) | 0.27 ± 0.01 | 0.23 ± 0.02 | 0.19 ± 0.01 | 0.16 ± 0.01 * | 1.68 |
| Root dry weight (g·plant−1) | 0.03 ± 0.002 | 0.03 ± 0.001 | 0.02 ± 0.001 | 0.02 ± 0.001 ** | 1.5 |
| Accession | Description | Gene | Control | Treated |
|---|---|---|---|---|
| Photosynthesis | ||||
| tr|A7U3N0|A7U3N0_EUCGL | Ribulose bisphosphate carboxylase large chain (Fragment) | GN=rbcL PE=3 SV=1 | 7379.896 | 1713.233 |
| tr|D1MZ07|D1MZ07_EUCGL; tr|A0A059A414|A0A059A414_EUCGR; tr|A0A059AHP3|A0A059AHP3_EUCGR; tr|A0A059AI83|A0A059AI83_EUCGR; tr|A0A059AIQ7|A0A059AIQ7_EU | Ribulose bisphosphate carboxylase small chain | GN=EgRBCS2 PE=2 SV=1 | 13005.96 | 6464.88 |
| tr|A0A2N9QPM6|A0A2N9QPM6_9ROSA; tr|T1QP85|T1QP85 | Photosystem II D2 protein | GN=psbD PE=3 SV=1 | 31698.33 | 14556.42 |
| sp|Q49KW8|CYB6_EUCGG | Cytochrome b6 | GN=petB PE=3 SV=1 | 1029.958 | 550.3955 |
| Translation and transcription | ||||
| tr|A0A2N9QPN7|A0A2N9QPN7_9ROSA | 30S ribosomal protein S14_ chloroplastic | GN=rps14 PE=3 SV=1 | 827.5739 | 286.2062 |
| tr|A0A2K8GMY3|A0A2K8GMY3_9ROSA | 30S ribosomal protein S15_ chloroplastic | GN=rps15 PE=3 SV=1 | 366.9244 | 154.6282 |
| tr|A0A059AZP2|A0A059AZP2_EUCGR | Elongation factor Tu | GN=EUGRSUZ_H01524 PE=3 SV=1 | 433.0593 | 911.957 |
| tr|A0A059CKD9|A0A059CKD9_EUCGR | Eukaryotic translation initiation factor 5A | GN=EUGRSUZ_C00350 PE=3 SV=1 | 2096.565 | 1001.485 |
| tr|A0A059BE60|A0A059BE60_EUCGR | RNA-dependent RNA polymerase | GN=EUGRSUZ_G02093 PE=3 SV=1 | 2019.472 | 826.6111 |
| tr|A0A1S6XZH0|A0A1S6XZH0_EUCGL | C-repeat binding factor | GN=CBF1d PE=2 SV=1 | 2137.21 | 7867.328 |
| Antioxidant enzyme | ||||
| tr|A0A059B660|A0A059B660_EUCGR; tr|A0A059B6P5|A0A059B6P5_EUCGR; tr|A0A059B713|A0A059B713_EUCGR | Superoxide dismutase [Cu-Zn] | GN=EUGRSUZ_H04426 PE=3 SV=1 | 3380.004 | 9448.107 |
| tr|A0A059CT24|A0A059CT24_EUCGR | Peroxidase | GN=EUGRSUZ_C02744 PE=3 SV=1 | 1737.219 | 751.3391 |
| Metabolism process Energy and carbohydrate metabolism | ||||
| sp|Q49KZ1|ATPB_EUCGG | ATP synthase subunit beta_ chloroplastic | GN=atpB PE=3 SV=1 | 51102.71 | 33223.95 |
| tr|A0A2K8GMV7|A0A2K8GMV7_9ROSA | ATP synthase subunit | GN=atpB PE=3 SV=1 | 23006.91 | |
| sp|Q49KZ2|ATPE_EUCGG | ATP synthase epsilon chain_ chloroplastic | GN=atpE PE=3 SV=1 | 7304.724 | 4168.356 |
| Glycolysis and carbohydrate metabolism | ||||
| tr|A0A059B8M0|A0A059B8M0_EUCGR | Glyceraldehyde-3-phosphate dehydrogenase (Fragment) | GN=EUGRSUZ_H04673 PE=3 SV=1 | 1885.399 | 5275.84 |
| tr|A0A059A4U5|A0A059A4U5_EUCGR; tr|A0A059A3P2|A0A059A3P2_EUCGR | Fructose-bisphosphate aldolase | GN=EUGRSUZ_K02073 PE=3 SV=1 | 2140.208 | 4374.579 |
| tr|A0A059C2Y2|A0A059C2Y2_EUCGR; tr|A0A059C3I0|A0A059C3I0_EUCGR | Phosphoribulokinase | GN=EUGRSUZ_E01261 PE=3 SV=1 | 62215.04 | 30478.44 |
| tr|I0IK58|I0IK58_9MYRT; tr|I0IK59|I0IK59_9MYRT | Sucrose synthase (Fragment) | GN=SuSy1 PE=3 SV=1 | 632.5789 | 1286.607 |
| tr|A0A059C344|A0A059C344_EUCGR | Starch synthase_ chloroplastic/amyloplastic | GN=EUGRSUZ_E01068 PE=3 SV=1 | 1051.751 | 294.2494 |
| tr|A0A059BWS2|A0A059BWS2_EUCGR; tr|A0A059BX43|A0A059BX43_EUCGR | Phospholipase D | GN=EUGRSUZ_F03862 PE=3 SV=1 | 439.2654 | 200.1259 |
| tr|A0A059BV12|A0A059BV12_EUCGR | Malate dehydrogenase | GN=EUGRSUZ_F03251 PE=3 SV=1 | 947.0006 | 2298.791 |
| sp|P46487|MDHM_EUCGU | Malate dehydrogenase_ mitochondrial | GN=MDH PE=2 SV=1 | 6368.574 | 2733.391 |
| Unknown | ||||
| tr|A0A059DFL1|A0A059DFL1_EUCGR | Purple acid phosphatase | GN=EUGRSUZ_A01512 PE=3 SV=1 | 7115.349 | 15029.53 |
| tr|A0A059BJT7|A0A059BJT7_EUCGR | Patatin | GN=EUGRSUZ_F00259 PE=3 SV=1 | 14.78243 | 618.4795 |
| tr|A0A059BV73|A0A059BV73_EUCGR | Probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 | GN=EUGRSUZ_F03307 PE=3 SV=1 | 2655.225 | 777.6948 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, M.O.; Mohammed, A.E.; Almutairi, T.A.; Elobeid, M.M. RETRACTED: Morpho-Physiological and Proteomic Analyses of Eucalyptus camaldulensis as a Bioremediator in Copper-Polluted Soil in Saudi Arabia. Plants 2019, 8, 43. https://doi.org/10.3390/plants8020043
Alotaibi MO, Mohammed AE, Almutairi TA, Elobeid MM. RETRACTED: Morpho-Physiological and Proteomic Analyses of Eucalyptus camaldulensis as a Bioremediator in Copper-Polluted Soil in Saudi Arabia. Plants. 2019; 8(2):43. https://doi.org/10.3390/plants8020043
Chicago/Turabian StyleAlotaibi, Modhi O., Afrah E. Mohammed, Taghreed A. Almutairi, and Mudawi M. Elobeid. 2019. "RETRACTED: Morpho-Physiological and Proteomic Analyses of Eucalyptus camaldulensis as a Bioremediator in Copper-Polluted Soil in Saudi Arabia" Plants 8, no. 2: 43. https://doi.org/10.3390/plants8020043





