Insertional Mutagenesis Approaches and Their Use in Rice for Functional Genomics
Abstract
:1. Introduction
2. Insertional Mutagenesis
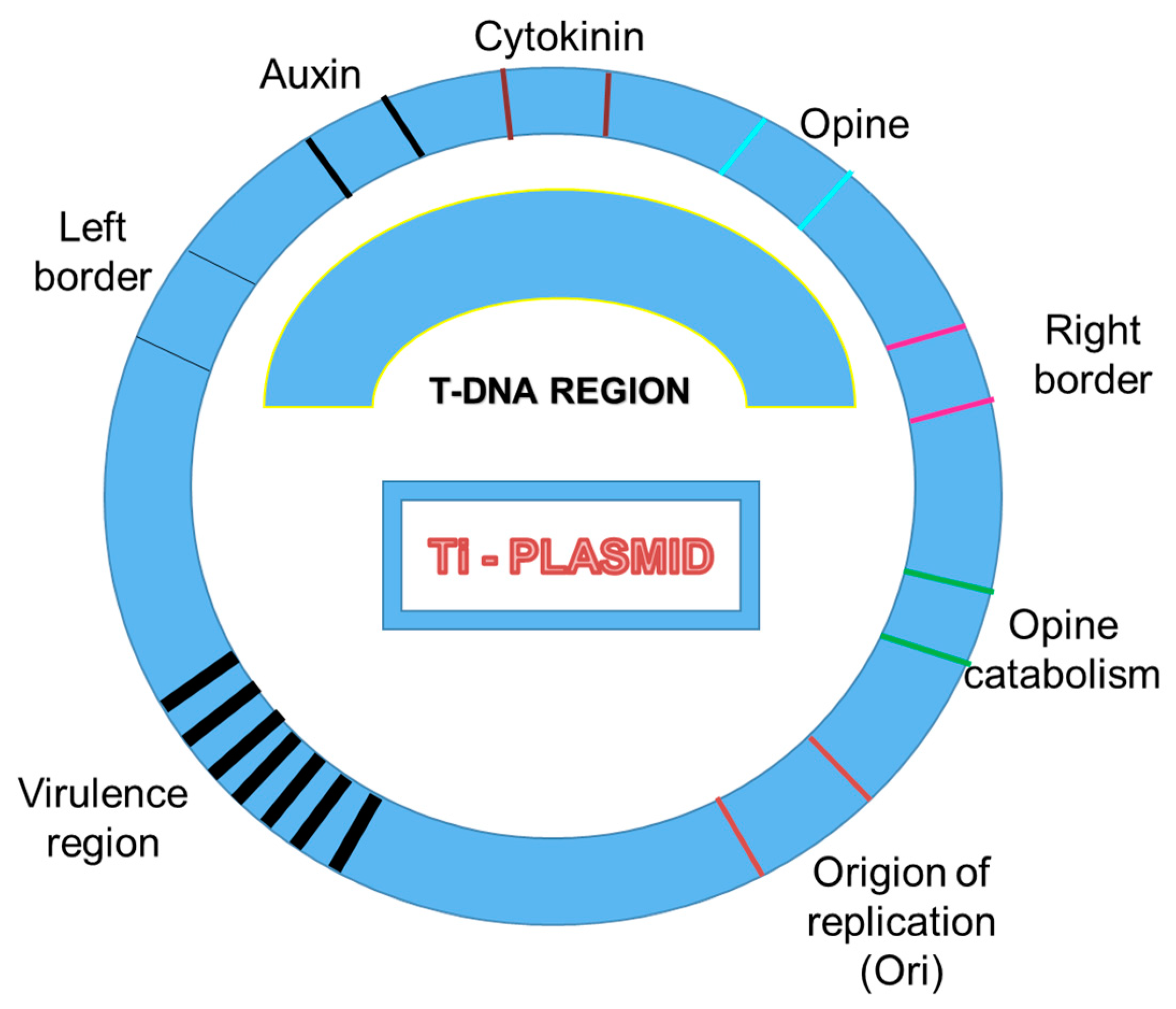
3. T-DNA Insertional Mutagenesis
4. Transposon Integration Mutagenesis
5. Entrapment Tagging
6. Screening of Insertional Mutants
7. Rice T-DNA Mutant Databases and Tools
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [PubMed]
- Kodym, A.; Afza, R. Physical and chemical mutagenesis. Methods Mol. Biol. 2003, 236, 189–204. [Google Scholar] [PubMed]
- Wu, C.; Li, X.; Yuan, W.; Chen, G.; Kilian, A.; Li, J.; Xu, C.; Li, X.; Zhou, D.X.; Wang, S.; et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003, 35, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M. Induced Mutagenesis in Plants Using Physical and Chemical Agents. In Plant Cell Culture: Essential Methods; Wiley: Hoboken, NJ, USA, 2010; Volume 20, pp. 111–130. ISBN 9780470686485. [Google Scholar]
- Krishnan, A.; Guiderdoni, E.; An, G.; Hsing, Y.C.; Han, C.D.; Lee, M.C.; Yu, S.M.; Upadhyaya, N.; Ramachandran, S.; Zhang, Q.; et al. Mutant Resources in Rice for Functional Genomics of the Grasses. PLANT Physiol. 2009, 149, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maarse, H. Rice. In Volatile Compounds in Foods and Beverages; Springer: New Delhi, India, 2017; pp. 79–89. ISBN 9781351405355. [Google Scholar]
- Lawrence, C.W. Classical mutagenesis techniques. Methods Enzymol. 1991, 194, 273–281. [Google Scholar] [PubMed]
- Krieg, D.R. Ethyl Methanesulfonate-Induced Reversion Of Bacteriophage T4rii Mutants. Genetics 1963, 48, 561. [Google Scholar] [PubMed]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R.; et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Henikoff, S. TILLING: Practical single-nucleotide mutation discovery. Plant J. 2006, 45, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, A.; Gupta, N.K.; Raj Jain, N.; Nayama, S. Studies on the Effect of Plant Growth Regulators and Micronutrients on Okra (Abelmoschus esculentus L) cv. Parbhani Kranti. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 3216–3223. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, C.K.; Kang, M.; Ji, S.U.; Yoon, U.H.; Kim, Y.H.; Lee, G.S. A Gene Functional Study of Rice Using Ac/Ds Insertional Mutant Population. Plant Breed. Biotech. 2018, 6, 313–320. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Cas9/sgRNA-based genome editing and other reverse genetic approaches for functional genomic studies in rice. Brief. Funct. Genom. 2018, 17, 339–351. [Google Scholar] [CrossRef]
- Springer, P.S. Gene traps: Tools for plant development and genomics. Plant Cell 2000, 12, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-L.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.R.S.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.J.; et al. Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.-H.; Yu, M.; Lai, E.M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhou, D.X. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. USA 2008, 105, 13679–13684. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Sparks, C.; Amoah, B.; Jones, H.D. Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep. 2003, 21, 659–668. [Google Scholar] [PubMed]
- Dubin, M.J.; Mittelsten Scheid, O.; Becker, C. Transposons: A blessing curse. Curr. Opin. Plant Biol. 2018, 42, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Babakhani, S.; Oloomi, M. Transposons: The agents of antibiotic resistance in bacteria. J. Basic Microbiol. 2018, 58, 905–917. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Lee, S.; Kim, S.H.; Kim, S.R. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005, 46, 14–22. [Google Scholar] [CrossRef]
- Munoz-Lopez, M.; Garcia-Perez, J. DNA Transposons: Nature and Applications in Genomics. Curr. Genom. 2010, 11, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Kolesnik, T.; Szeverenyi, I.; Bachmann, D.; Kumar, C.S.; Jiang, S.; Ramamoorthy, R.; Cai, M.; Ma, Z.G.; Sundaresan, V.; Ramachandran, S. Establishing an efficient Ac/Ds tagging system in rice: Large-scale analysis of Ds flanking sequences. Plant J. 2004, 37, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Ramachandran, S. Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PLoS ONE 2011, 6, e28069. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Bao, Z.; Zhang, X.; Hirochika, H.; Eddy, S.R.; McCouch, S.R.; Wessler, S.R. An active DNA transposon family in rice. Nature 2003, 421, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Terauchit, K.; Wada, M.; Hirano, H.Y. The plant MITE mPing is mobilized in anther culture. Nature 2003, 421, 167–170. [Google Scholar] [CrossRef]
- Nakazaki, T.; Okumoto, Y.; Horibata, A.; Yamahira, S.; Teraishi, M.; Nishida, H.; Inoue, H.; Tanisaka, T. Mobilization of a transposon in the rice genome. Nature 2003, 421, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Hancock, C.N.; Zhang, F.; Floyd, K.; Richardson, A.O.; LaFayette, P.; Tucker, D.; Wessler, S.R.; Parrott, W.A. The Rice Miniature Inverted Repeat Transposable Element mPing Is an Effective Insertional Mutagen in Soybean. Plant Physiol. 2011, 157, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, S.; Tsukiyama, T.; Okumoto, Y.; Tanisaka, T. Early Embryogenesis-Specific Expression of the Rice Transposon Ping Enhances Amplification of the MITE mPing. PLoS Genet. 2014, 10, e1004396. [Google Scholar] [CrossRef] [PubMed]
- Ram, H.; Kaur, A.; Gandass, N.; Singh, S.; Deshmukh, R.; Sonah, H.; Sharma, T.R. Molecular characterization and expression dynamics of MTP genes under various spatiotemporal stages and metal stress conditions in rice. PLoS ONE 2019, 14, e0217360. [Google Scholar] [CrossRef] [PubMed]
- Hirochika, H. Retrotransposons of rice: Their regulation and use for genome analysis. Plant Mol. Biol. 1997, 35, 231–240. [Google Scholar] [CrossRef]
- Chin, H.G.; Choe, M.S.; Lee, S.H.; Park, S.H.; Park, S.H.; Koo, J.C.; Kim, N.Y.; Lee, J.J.; Oh, B.G.; Yi, G.H.; et al. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 1999, 19, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.-H.; Hur, J.; Ryu, C.-H.; Choi, Y.; Chung, Y.-Y.; Miyao, A.; Hirochika, H.; An, G. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Paszkowski, J. Plant genome modification by homologous recombination. Curr. Opin. Plant Biol. 2003, 6, 157–162. [Google Scholar] [CrossRef]
- Feldmann, K.A. T—DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1991, 1, 71–82. [Google Scholar] [CrossRef]
- Jeon, J.S.; Lee, S.; Jung, K.H.; Jun, S.H.; Jeong, D.H.; Lee, J.; Kim, C.; Jang, S.; Lee, S.; Yang, K.; et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000, 22, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.H.; You, J.H.; Kang, H.G.; Hur, J.; Kim, Y.H.; Han, M.J.; An, K.; Chung, B.C.; Lee, C.H.; An, G. Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol. Biol. 2004, 54, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; An, S.; Park, S.; Kang, H.G.; Park, G.G.; Kim, S.R.; Sim, J.; Kim, Y.O.; Kim, M.K.; Kim, S.R.; et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006, 45, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2005, 34, D745–D748. [Google Scholar] [CrossRef]
- Hsing, Y.I.; Chern, C.G.; Fan, M.J.; Lu, P.C.; Chen, K.T.; Lo, S.F.; Sun, P.K.; Ho, S.L.; Lee, K.W.; Wang, Y.C.; et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007, 63, 351–364. [Google Scholar] [CrossRef]
- Sallaud, C.; Gay, C.; Larmande, P.; Bès, M.; Piffanelli, P.; Piégu, B.; Droc, G.; Regad, F.; Bourgeois, E.; Meynard, D.; et al. High throughput T-DNA insertion mutagenesis in rice: A first step towards in silico reverse genetics. Plant J. 2004, 39, 450–464. [Google Scholar] [CrossRef]
- Johnson, A.A.T.; Hibberd, J.M.; Gay, C.; Essah, P.A.; Haseloff, J.; Tester, M.; Guiderdoni, E. Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J. 2005, 41, 779–789. [Google Scholar] [CrossRef]
- Van Enckevort, L.J.G.; Droc, G.; Piffanelli, P.; Greco, R.; Gagneur, C.; Weber, C.; González, V.M.; Cabot, P.; Fornara, F.; Berri, S.; et al. EU-OSTID: A collection of transposon insertional mutants for functional genomics in rice. Plant Mol. Biol. 2005, 59, 99–110. [Google Scholar] [CrossRef]
- Wan, S.; Wu, J.; Zhang, Z.; Sun, X.; Lv, Y.; Gao, C.; Ning, Y.; Ma, J.; Guo, Y.; Zhang, Q.; et al. Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 2009, 69, 69–80. [Google Scholar] [CrossRef]
- Krysan, P.J.; Young, J.C.; Sussman, M.R. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 1999, 11, 2283–2290. [Google Scholar] [CrossRef]
- Yao, W.; Li, G.; Yu, Y.; Ouyang, Y. FunRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 2018, 7, gix119. [Google Scholar] [CrossRef]
- Jeong, D.-H. T-DNA Insertional Mutagenesis for Activation Tagging in Rice. Plant Physiol. 2002, 130, 1636–1644. [Google Scholar] [CrossRef] [Green Version]
- Miyao, A. Target Site Specificity of the Tos17 Retrotransposon Shows a Preference for Insertion within Genes and against Insertion in Retrotransposon-Rich Regions of the Genome. Plant Cell 2003, 15, 1771–1780. [Google Scholar] [CrossRef]
- Upadhyaya, N.M.; Zhu, Q.H.; Zhou, X.R.; Eamens, A.L.; Hoque, M.S.; Ramm, K.; Shivakkumar, R.; Smith, K.F.; Pan, S.T.; Li, S.; et al. Dissociation (Ds) constructs, mapped Ds launch pads and a transiently-expressed transposase system suitable for localized insertional mutagenesis in rice. Theor. Appl. Genet. 2006, 112, 1326–1341. [Google Scholar] [CrossRef]
- Kumar, C.S.; Wing, R.A.; Sundaresan, V. Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J. 2005, 44, 879–892. [Google Scholar] [CrossRef]
- Droc, G.; Périn, C.; Fromentin, S.; Larmande, P. OryGenesDB 2008 update: Database interoperability for functional genomics of rice. Nucleic Acids Res. 2009, 37, D992–D995. [Google Scholar] [CrossRef]
- Droc, G.; An, G.; Wu, C.; Hsing, Y.C.; Hirochika, H.; Pereira, A.; Sundaresan, V.; Upadhyaya, N.; Ramachandran, S.; Comai, L.; et al. Mutant Resources for Functional Analysis of the Rice Genome. In Genetics and Genomics of Rice. Plant Genetics and Genomics: Crops and Models, 5th ed.; Zhang, Q., Wing, R.A., Eds.; Springer: New York, NY, USA, 2013; pp. 81–115. [Google Scholar]
- Wei, F.J.; Droc, G.; Guiderdoni, E.; Hsing, Y.I. International Consortium of Rice Mutagenesis: Resources and beyond. Rice (NY) 2013, 6, 39. [Google Scholar] [CrossRef]
- Wei, F.; Tsai, Y.; Hsu, Y.; Chen, Y.; Huang, C.; Wu, H.; Huang, L.; Lai, M.; Kuang, L.; Yu, S.; et al. Lack of genotype and phenotype correlation in a rice T-DNA tagged line is likely caused by introgression in the seed source. PLoS ONE 2016, 11, e0155768. [Google Scholar] [CrossRef]
- Chang, C.L.; Serapion, J.C.; Hung, H.H.; Lin, Y.C.; Tsai, Y.C.; Jane, W.N.; Chang, M.C.; Lai, M.H.; Hsing, Y.C. Studies of a rice sterile mutant sstl from the TRIM collection. Bot Stud. 2019, 60, 12. [Google Scholar] [CrossRef]
- Nakamura, H.; Hakata, M.; Amano, K.; Miyao, A.; Toki, N.; Kajikawa, M.; Pang, J.; Higashi, N.; Ando, S.; Toki, S.; et al. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 2007, 65, 357–371. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, T.; Kondou, Y.; Akiyama, K.; Kurotani, A.; Higuchi, M.; Ichikawa, T.; Kuroda, H.; Kusano, M.; Mori, M.; Saitou, T.; et al. RiceFOX: A database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011, 52, 265–273. [Google Scholar] [CrossRef]
- Meng, X.; Yu, H.; Zhang, Y.; Zhuang, F.; Song, X.; Gao, S.; Gao, C.; Li, J. Construction of a Genome-Wide Mutant Library in Rice Using CRISPR/Cas9. Mol. Plant 2017, 10, 1238–1241. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhu, J.K. Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 523–525. [Google Scholar] [CrossRef]


| Resource/Database Name | Institution | Genotype | Type of T-DNA Mutagen | Number of Available FSTs Lines a | Number of Mapped FSTs a | Website of the Resource/Database | Reference | Number of Citations b |
|---|---|---|---|---|---|---|---|---|
| POSTECH Rice Insertion Database (RISD) | Pohang University of Science and Technology, South Korea | Dongjin Hwayoung Kitaake | Gene trap, Activation tagging | 107,171 | 99,559 | http://cbi.khu.ac.kr/ | [36] [37] [38] | 684 94 452 308 |
| Rice Mutant Database (RMD) | Huazhong Agricultural University, China | Zhonghua 11 Zhonghua 15 Nipponbare | Enhancer trap, Tos17 | 85,315 | 91,792 | http://rmd.ncpgr.cn/ | [18] [39] | 251 183 |
| Taiwan Rice Insertion Mutant (TRIM) | Institute of Plant and Microbial Biology, Academia Sinica, Taiwan | Tainung 67 | Activation tagging | 59,804 | 58,764 | http://trim.sinica.edu.tw | [40] | 192 |
| Oryza Tag Line (OTL) Génoplante | CIRAD-INRA-IRD-CNRS, France | Nipponbare | Enhancer trap, Tos17 | 29,263 | 29,429 | http://oryzatagline.cirad.fr/ | [41] [42] | 268 104 |
| Shanghai T-DNA Insertion Population Database (SHIP) | Shanghai Institute of Plant Physiology and Ecology (SIPPE), China | Zhonghua 11 | Enhancer trap | 10,381 | 10,934 | http://www.plantsignal.cn/ | [43] | 42 |
| Chinese Academy of Agricultural Sciences (CAAS) | Beijing Biotechnology Research Institute, CAAS, China | Nipponbare | Activation tagging | N/A | N/A | N/A | [44] | 85 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ram, H.; Soni, P.; Salvi, P.; Gandass, N.; Sharma, A.; Kaur, A.; Sharma, T.R. Insertional Mutagenesis Approaches and Their Use in Rice for Functional Genomics. Plants 2019, 8, 310. https://doi.org/10.3390/plants8090310
Ram H, Soni P, Salvi P, Gandass N, Sharma A, Kaur A, Sharma TR. Insertional Mutagenesis Approaches and Their Use in Rice for Functional Genomics. Plants. 2019; 8(9):310. https://doi.org/10.3390/plants8090310
Chicago/Turabian StyleRam, Hasthi, Praveen Soni, Prafull Salvi, Nishu Gandass, Ankita Sharma, Amandeep Kaur, and Tilak Raj Sharma. 2019. "Insertional Mutagenesis Approaches and Their Use in Rice for Functional Genomics" Plants 8, no. 9: 310. https://doi.org/10.3390/plants8090310
APA StyleRam, H., Soni, P., Salvi, P., Gandass, N., Sharma, A., Kaur, A., & Sharma, T. R. (2019). Insertional Mutagenesis Approaches and Their Use in Rice for Functional Genomics. Plants, 8(9), 310. https://doi.org/10.3390/plants8090310






