1. Introduction
In modern horticulture, growers need to optimize plant development and yield [
1] in order to meet the food demand of increasing populations [
2], especially in developing countries [
3], and contribute towards food security and social stability. Within this scope, applied research on innovative horticultural practices can make effective use of dynamic crop growth models [
4] under conditions optimal for plant growth and for eliciting plant response to abiotic stresses [
5], therefore, allowing a more rational use of resources, such as water and nutrients [
4].
Several fundamental physiological processes such as photosynthesis, transpiration, and cooling are facilitated by leaves [
6] and they are, therefore, strongly influenced by leaf morphology (size, shape, symmetry, venation, organization, and petiole characteristics) [
7]. The characterization of leaf morphology and quantification of leaf area (LA) and/or leaf area index (LAI) is consequently of paramount importance to horticultural crop science. In this respect, there is an increasing interest in using computer-assisted imaging systems [
8] for producing reliable biometric measurements [
9] and analyzing phenotypic traits related to plant architecture and leaf characteristics [
10]. For instance, data on leaf characteristics can be incorporated into databases [
11,
12] and employed to validate time-series quantification of leaf morphology (e.g., [
13,
14]) and to determine the performance of computer-assisted imaging systems and machine learning algorithms used to classify/recognize phenotypic traits of specific genotypes [
15].
Leaf area is generally measured with destructive or non-destructive methods [
16], the latter often preferred as they are faster, cheaper, and non-invasive (i.e., no excision of leaves is required), therefore, permitting repeated and simultaneous measurements of LA and other physiological parameters (e.g., leaf gas exchange or fluorescence) on the same leaves.
Collected information, such as leaf blade length (L) and width (W) [
17,
18,
19,
20,
21,
22,
23,
24,
25] or the shape ratio of the leaf (L:W) [
26], can be useful for characterizing leaf functions and structure, based only on proxy variables. In particular, the leaf shape ratio is of particular importance in horticultural sciences as it is regulated by several genetic factors and mutations [
27], whose diversity can be analyzed in functional [
28] and evolutionary terms [
29].
Thus far, numerous models have been proposed and applied with respect to both leaf (e.g., [
20,
30,
31]) and shoot level [
31,
32,
33,
34,
35,
36,
37,
38,
39,
40,
41] morphology of several fruit, vegetable, ornamental, medicinal, and aromatic crops [
42]. Currently, LA models for aromatic and medicinal plants comprise several species such as basil, winter red Bergenia, or purple bergenia, calamint, coffee, cherry laurel, bush-willows, jimson weed, wild cucumber, horse-eye bean, lemon balm, peppermint, oleander, mountain mint, opium poppy, ground-cherry, or winter cherry, picrorhiza or kutka, saffron, sugar leaf, snowbell, summer snowflake, tea, common nettle, orange mullein [
42], valeriana [
43], and pepper plants [
44].
The world production of medicinal and aromatic plants (MAPs) is expected to rise up to 5 trillion US
$ by 2050 [
45]. Thanks to their aromatic oils [
45] and other phytochemical constituents MAPs are used (a) to deter herbivores, pathogens, and parasites, (b) as culinary herbs and spices (e.g., thyme, laurel, and basil), (c) to produce scent and (d) as ornamentals (e.g.,
Eucalyptus spp.,
Lavandula spp., and
Cistus spp.). In this context, there is an increasing interest in collecting systematic information on the physiology and phenotypic traits of these crops, information that can be used, for instance, to simulate seasonal variations of leaf area and, therefore, estimate periodic treatments and the needs for irrigation during the cultivation of MAPs. In addition, precise estimations of LA will be necessary to develop more complex process-based plant growth models that can be used to build support decision systems to help growers in managing fertilization and irrigation. Therefore, the aims of this work were to employ rigorous statistical analysis in order to (1) test and compare fast but accurate generalized allometric models for different cultivars of basil, mint, and sage—three well-common MAPs used worldwide as culinary herbs—and (2) validate the best models using an independent dataset.
3. Discussion
The importance of this study lies in the fact that the leaf and, by extrapolation, the entire canopy represent the fundamental physiological hubs of photosynthesis and of gas exchange with the atmosphere. As the ability to intercept light is clearly dependent on the two-dimensional leaf structure (i.e., shape and area) [
27], the characterization of leaf L and W in several species has a large value within the broad fields of botany, plant physiology, and crop science. This information can be also needed in the future to determine the performance of novel photogrammetry and computer vision algorithms used to characterize whole-plant phenotypic traits of specific genotypes [
15]. In this study, species-specific data on phenotypic traits (leaf L and W) of basil, mint, and sage were collected and, following rigorous statistical analysis, they were used to calibrate and validate five single leaf area (LA) allometric models.
To be effective, a single leaf model needs to be accurate over a large range of variability of phenotypic traits, a condition that is attainable by (a) using data for calibration collected on a large number of cultivars, and (b) validating the final model using additional independent dataset. For this reason, we gave particular importance to the preliminary characterization of phenotypic traits by using three different species and 14 cultivars (5, 4, and 5 cultivars for basil, mint, and sage, respectively), and by choosing three additional cultivars to validate the best ranking models. As reported by Tsukaya [
28], leaf W and narrow leaf shape (larger leaf index = stenophylly) might be driven evolutionarily by the growth habitat and other external conditions experienced by a species. In this context, many factors and different genes are thought to be involved in the selection of a particular leaf W for any given species and, therefore, as a final result, in the evolution of the leaf shape index [
28]. All the species analyzed in this study belong to the
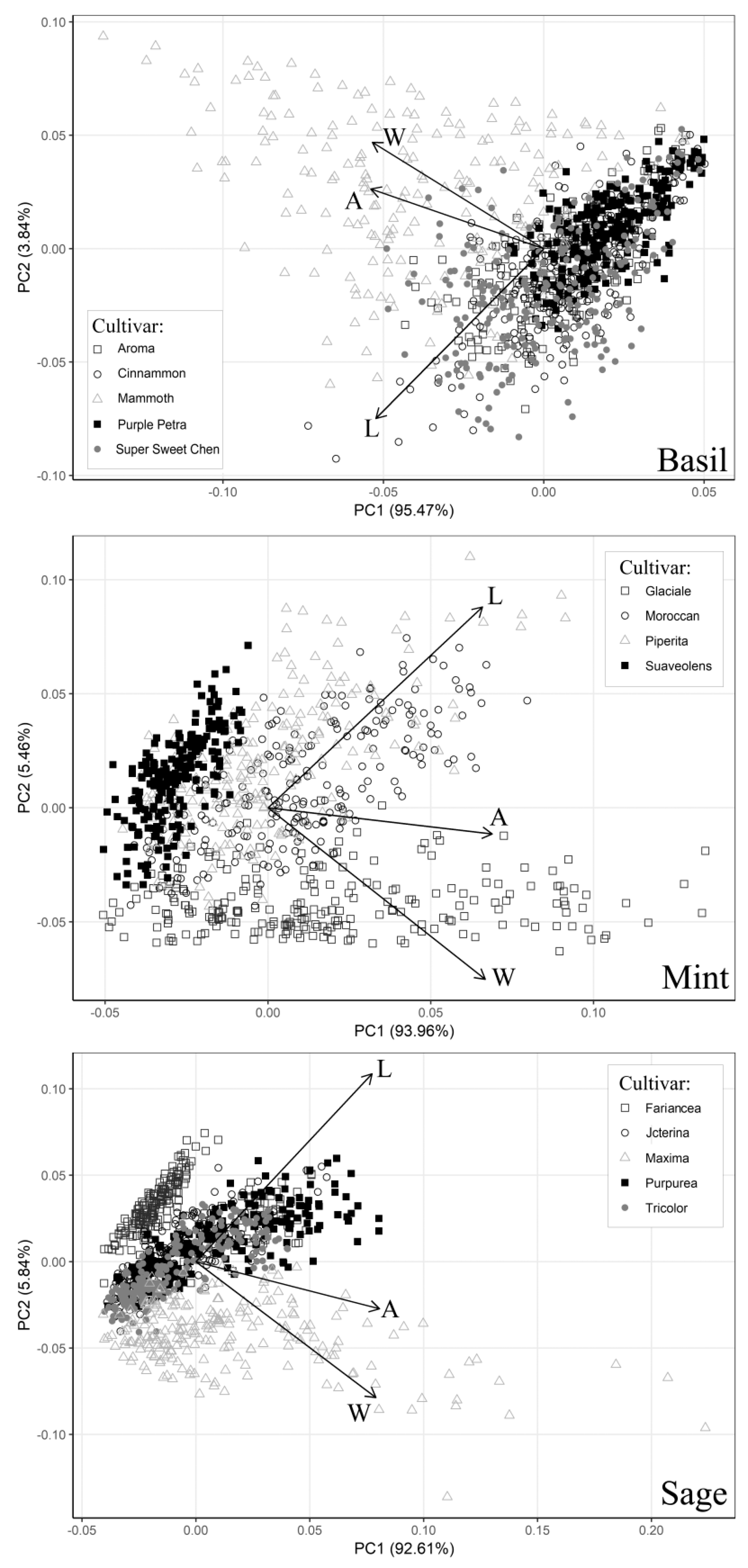
Lamiaceae, a family having as the main center of variability the Mediterranean basin generally in degraded areas such as maquis and garrigues with rocky, calcareous, or sandy soils. In this case, the leaf shape ratio estimated on pooled data (calibration sets;
Table 6) showed that sage had a narrow leaf (L:W = 2.4) and the largest variability, whereas basil and mint cultivars showed a quite similar leaf shape ratio with a mean value of about 1.5. At the same time, the PCA highlighted the existence, in some cultivars, of a good correlation between leaf W and single LA, thus explaining the better fitting and higher accuracy showed in some cases by the quadratic model based on the squared leaf W parameter (model no. 5).
The PCA also highlighted, for some cultivars, a good correlation (especially in mint) between the single LA and leaf L. These results were consistent with what observed graphically using PC1 and PC2. Indeed, some cultivars were distributed principally along with the leaf L loading, whereas others were either associated with the leaf W loading or mainly concentrated near the origin of the axes (
Figure 1).
Comparing the PCA graphs with the ability of the five models to predict the single LA of the calibration set cultivars, it is clear that the observed intra-specific variability reduced the effectiveness of the one-regressor models. Moreover, during the validation phase, the one-regressor model no. 5 performed moderately well only for basil, but when it was used for mint and sage, blade size of small and large leaves was overestimated and underestimated, respectively. Indeed, the residual plots (
Figures S2A and S3A) confirmed that model no. 5 was not a good fit, at least for mint and sage species. Although the results showed the limitation of models based on a fast single measurement (in particular in mint and sage) at the expense of a slightly increased variation, one-regressor models based on leaf W can be used in calibrated cultivars, especially if a range of different growing conditions (e.g., open-field and greenhouse) and situations are considered and included during the calibration and validation phases. This is also consistent with previous results reported by Gao et al. [
9] for rose genotypes and by Teobaldelli et al. [
46] for Loquat. On the contrary, a single leaf area could be estimated using the product of L and W without calibration per genotype, as reported by Rouphael et al. [
30] and Teobaldelli et al. [
46]. All these important aspects were also confirmed in our study for the three analyzed MAPs species. Indeed, our results outlined the importance of the two-regressors model (no. 3) based on the product of L × W, that was able to provide accurate predictions of the single LA both for all the three species and for each cultivar during the calibration and validation phases. This result is consistent also with previous studies on LA estimation in several fruits and horticultural crops ([
18,
36,
47,
48,
49]).
Our study was carried out using healthy leaves collected on unstressed plants grown in a greenhouse. Thus, it cannot be excluded that the models developed in our study will need to also be validated under other growing conditions and under different intensity levels of biotic and abiotic stresses. However, previous studies [
50] carried out on eggplants reported that models calibrated using cultivars grown under open-field conditions might provide also good results if validated against data collected in greenhouses.
4. Materials and Methods
4.1. Growth Conditions, Plant Material, and Data Collection
A greenhouse experiment was carried out during the 2011 spring at the private farm Gli Aromi (36°45′42.50″ N 14°42′36.89″ E) located in Sicily, southern Italy. The tested crops for the present study were 3 medicinal and aromatic plants (MAPs) species, namely basil (Ocimum basilicum L.), mint (Mentha spp.), and sage (Salvia spp.). Plants were grown according to the same commercial protocol and under identical natural light conditions. Inside the greenhouse, the mean air temperature was 24 °C, varying between 19 and 31 °C and the relative humidity was 60% and 75% during day and night, respectively. The tested medicinal and aromatic plants were grown in plastic containers (diameter: 14 cm; height: 12 cm) containing a peat/perlite mixture in a 1:1 volume ratio.
For model calibration, the trial included a total of 5 basil cultivars (‘Aroma’, ‘Cinnamon’, ‘Mammoth’, ‘Purple Petra’ and ‘Super Sweet Chen’; four mint cultivars (‘Moroccan’ [Mentha spicata L.], ‘Piperita’ [Mentha × piperita L.], ‘Glaciale’ [Mentha × rotundifolia (L.) Huds. and ‘Suaveolens’ [Mentha suaveolens Ehrh.]); and five sage cultivars (‘Jcterina’, ‘Tricolor’ and ‘Maxima’ [Salvia officinalis L.], ‘Purpurea’ [Salvia purpurea Cav.] and ‘Fariancea’ [Salvia farinacea Benth.]). For model validation, the ‘Lettuce Leaf’, ‘Comune’ and also ‘Comune’ cultivars were used for basil, mint, and sage, respectively. These cultivars were selected as representatives of the basil, mint, and sage cultivated in the south Mediterranean region including Italy, Spain, and Greece.
A total of 206 to 235 healthy leaves were collected for each of the 14 cultivars used for model calibration, whereas 400 to 500 healthy leaves were collected for each of the 3 cultivars used for model validation. Leaves with a minimum blade width of 0.5 cm were randomly collected in spring from different levels of the canopy in order to capture the natural variability in the leaf shape of each cultivar. Collected leaves were rapidly transported to the laboratory where the parameters L, W, and LA of the leaf blades were individually measured (
Figure 8). LA was measured with an LA-meter (LI-3100; LICOR, Lincoln, NE, USA) calibrated to 0.01 cm
2.
4.2. Statistical Analysis
All the measurements were carried out using R-STAT (a free software environment for statistical computing and graphics; version n. 3.5.2, release codenamed “Eggshell Igloo”; © 2018 of The R Foundation for Statistical Computing) and packages available in the CRAN (comprehensive R archive network) repository [
51].
4.2.1. Principal Component Analysis
To understand how different phenotypic traits such as leaf L and W influenced the single LA of different cultivars within each species, the Pearson correlation between the three parameters was estimated using the cor() function of the ‘Stats’ package [
51]. Moreover, a principal component analysis (PCA), based on the singular value decomposition (SVD) [
52], was carried out following the indications of Manly and Alberto [
53], using the prcomp() function, also available in the ‘Stats’ package [
51].
4.2.2. Model Calibration
Five different allometric models (3 linear and 2 quadratic) were chosen based on leaf phenotypic traits [
26] of the 3 selected species, and were used to estimate the single LA based on fast measurements of 2 proxy variables as follows:
where
a and
b are the coefficients of the linear and quadratic models, LA is the single leaf area (cm
2), L is the leaf length (cm), and W is the leaf width (cm).
Regarding basil, LA estimation was carried out by aggregating data (calibration set,
n = 1094;
Table 6) measured on 5 cultivars, i.e., ‘Aroma’, ‘Cinnamon’, ‘Mammoth’, ‘Purple Petra’ and ‘Super Sweet Chen’. In the case of mint, the pooled data (calibration set,
n = 901;
Table 6) were collected on 4 cultivars, namely ‘Glaciale’, ‘Moroccan’, ‘Piperita’ and ‘Suaveolens’. The pooled calibration dataset (
n = 1103;
Table 6) for sage was based on data collected on 5 cultivars, i.e., ‘Fariancea’, ‘Jcterina’, ‘Maxima’, ‘Purpurea’ and ‘Tricolor’.
The linear and quadratic models were fitted using the lm() function of the ‘Stats’ package [
51]. The following criteria were used to evaluate the performance of the 5 allometric models: R-square (R²), root mean square error (RMSE in cm
2) and the Bayesian information criterion (BIC) [
54]. The BIC() function, available in the ‘Stats’ [
51] package was used to calculate the BIC criterion. Finally, the best ranking model, both for the one-regressor and two-regressors models, was chosen as that having the highest R
2 and the lowest RMSE and BIC. Therefore, the coefficients of the selected models were optimized using a non-parametric bootstrap function available on the ‘Boot’ package [
55]. The coefficients of the bootstrap model together with the associated standard, median, and percentage confidence intervals were obtained iteratively in 1000 selected bootstrap samples with substitution of observations from the original data set.
4.2.3. Model Validation
The selected bootstrapped models (with one or double predictors) were validated by comparing the predicted single leaf area (PLA) values estimated using L and/or W values, obtained during the validation experiment with the observed single LA (OLA) values of the following cultivars:
Cultivar ‘Lettuce Leaf’ (n = 487) for basil;
Cultivar ‘Comune’ (n = 441) for mint;
Cultivar ‘Comune’ (n = 418) for sage.
The goodness of estimation of the selected models was evaluated by analyzing the R² and the RMSE (cm2) of the observed leaf area (OLA) compared to the predicted leaf area (PLA) for each cultivar used to validate each model.
5. Conclusions
Much research has been conducted on several fruit crops and MAPs to characterize leaf traits. In addition, numerous models based generally on two important proxy parameters, such as leaf L and/or W have been proposed, tested, and validated. In this study, single LA of basil, mint and sage, was predicted using five allometric models (linear or quadratic) based both on leaf L or W or on the product of these two variables. Our results confirmed the capability of two-regressors models, especially when intra-specific variability is considered, to estimate single LA for the abovementioned species. Indeed, this study suggested that by using a two-regressors model based on the product of leaf W and L, predictions can be quite accurate without the need to calibrate model coefficients for genotype. Nevertheless, calibration might be required if the selected cultivars are growing under biotic and/or abiotic stress conditions.
On the other hand, even if a good fitting can be found using the one-regressor model based on squared leaf W (model no. 5), predictions (especially in the case of mint and sage) might be biased.
Eventually, estimates of single LA of basil, mint, and sage, obtained with different levels of accuracy using one or more parameters and the validated coefficients can be a valid resource for plant physiologists, horticulturists, and growers to better estimate the single leaf area and the seasonal variation of LAI of their crops, to parameterize models (e.g., to estimate evapotranspiration) or to extract general information regarding the vigor and the growth stage of their crops. Since two-regressors models allowed very accurate estimations of leaf blade size of basil, sage, and mint, they can be useful also to support plant physiology studies and to develop more complex process-based plant growth models that can be used to build support decision systems to help growers in managing fertilization and irrigation.