Abstract
Although the nitrate assimilation into amino acids in photosynthetic leaf tissues is active under the light, the studies during 1950s and 1970s in the dark nitrate assimilation provided fragmental and variable activities, and the mechanism of reductant supply to nitrate assimilation in darkness remained unclear. 15N tracing experiments unraveled the assimilatory mechanism of nitrogen from nitrate into amino acids in the light and in darkness by the reactions of nitrate and nitrite reductases, glutamine synthetase, glutamate synthase, aspartate aminotransferase, and asparagine synthetase. Nitrogen assimilation in illuminated leaves and non-photosynthetic roots occurs either in the redundant way or in the specific manner regarding the isoforms of nitrogen assimilatory enzymes in their cellular compartments. The electron supplying systems necessary to the enzymatic reactions share in part a similar electron donor system at the expense of carbohydrates in both leaves and roots, but also distinct reducing systems regarding the reactions of Fd-nitrite reductase and Fd-glutamate synthase in the photosynthetic and non-photosynthetic organs.
1. Introduction
Plants use inorganic nitrogen present in the soil for their growth mainly in the form of nitrate (NO3−). Following the absorption through the roots, the most oxidized form of NO3− (+6) is reduced to organic forms (–2) such as amino acids prior to their incorporation into proteins, nucleotides, and chlorophylls. Although plants can assimilate NO3− to amino acids in both the photosynthetic leaves and non-photosynthetic roots, the major sites of nitrate assimilation are green shoots where energy (ATP), reductant (electrons) and organic skeletons are produced by photosynthetic reactions using solar energy. In addition, the nitrate assimilation in the leaves takes place in the night, i.e., by using storage carbohydrates [1]. However, the activities of nitrate assimilation in darkness measured from the 1950s to 1970s were fragmental and variable. Here, we present physiological aspects of light-independent nitrate-to-asparagine assimilation in the leaves by referring to the light-independent nitrate assimilation in the non-photosynthetic roots at the expense of carbohydrates, supplied by transport from the shoots, as reviewed recently [2].
2. 15N Tracing Analysis of Dark Nitrate Assimilation into Amino Acids in Leaves
Direct evidence of dark nitrate assimilation into amino acids in the leaves was obtained by incubating the leaf tissues with 15N-nitrate in darkness. Delwiche [3] fed detached immature leaves of tobacco (Nicotiana tabacum) with 15N-KNO3 solution (15 atom % excess 15N) for 24 h via petioles in darkness, and detected 15N-labelling in ammonia plus amide-N, and in ammonia fraction by 2.89 and 0.61 atom % excess 15N, respectively. Mendel and Visser [4] conducted a short incubation (30 min) of tomato leaf discs with 15N-KNO3 (14 atom % excess 15N) in the dark, and detected 15N-labelling in free ammonia fraction in duplicated samples by 2.13 and 2.42 atom % excess 15N. Canvin and Atkins [5] incubated leaf segments of wheat (Triticum aestivum L.) and corn (Zea mays L.) with 15N-NaNO3 solution (95 atom % 15N) for 15 and 30 min in darkness. After 15 min, they detected little 15N enrichment in the soluble amino acid fraction in darkness compared with the high 15N labelled amino acids in the light and concluded that the nitrate assimilation was strictly dependent on the light.
Shortly after the findings of 15N-labelled amino acids in the roots of rice (Oryza sativa L.) seedlings by feeding with 15N-ammonium [6] and 15N-nitrate [7], Ito and Kumazawa [8] conducted light and dark incubation of leaf discs of sunflower (Helianthus annuus L.) with 15NO3−, 15NO2−, and 15NH4+ for 30 min. They detected 15N-labelled glutamine (amino-N and amide-N), glutamate, and aspartate as shown in Table 1. In these studies, 15N-labelling of individual amino acids was determined by a combination of their separation on two-dimension thin-layer chromatography and 15N enrichment by emission optical spectrometry, as first described by Yoneyama and Kumazawa [6]. Dark 15N-labelling in the amino-N of glutamine, glutamate, and aspartate was less than that in the light, while the dark 15N-labelling of the amide-N of glutamine was higher than that in the light irrespective of the feeding with 15NO3−, 15NO2−, or 15NH4+. The results indicated that the transfer rate of the glutamine amide-N to 2-OG forming glutamate in darkness was lower than in the light although nitrate was reduced sequentially to nitrite and ammonia in darkness. It is noteworthy that in the light, the 15N-labellings of glutamine amide-N from all of 15NO3−, 15NO2−, and 15NH4+ were apparently lower comparing with those in darkness. This can be explained by a pool of NH4+ diluted by a large amount of photorespiratory NH4+ [9] and assimilated into glutamine amide-N by glutamine synthetase (Table 1). The non-photosynthetic root tissues, which have no photorespiratory activity, actively assimilated 15N into 15N-amide of glutamine and glutamate by the root feeding with 15NO3− or 15NO2− [2] as observed in the leaves in darkness (Table 1).

Table 1.
The 15N atom % excess of amino acids extracted from the sunflower leaf discus treated with 1 mM 15NO3−, 15NO2−, and 15NH4+ for 30 min under darkness as compared to those under the light (20,000 lux).
In the 1970s, a simple in vivo assay of nitrate reductase activity was widely employed using leaf segments without extraction of enzymes. In this technique, it was assumed that nitrite, nitrate reductase reaction product, was barely assimilated when the assay was carried out in the dark [10,11,12]. In vivo nitrate reductase assays were conducted using 15N-NaNO3 to measure its reduction to nitrite and the assimilation into amino acids under aerobic or anaerobic conditions [13]. Nitrite production was active under anaerobic conditions while 15N incorporation into amino acids was intensive under aerobic conditions than under anaerobic conditions (Table 2).

Table 2.
Nitrite formation and 15N incorporation into amino acids in the wheat leaf segments treated with 50 mM Na15NO3− for 60 min under aerobic (in air) or anaerobic (in N2) conditions in darkness.
The 15N-labelling experiments were carried out using green and white chlorophyll-less leaves of albino mutant seedlings of rice, which were produced by chemical mutation. It was shown that the white leaves fed with 15NO2− in darkness [14] had lower 15N-enrichments (atom % excess 15N) in glutamine, glutamate, and aspartate than green leaves, but a large 15N accumulation occurred in glutamine and particularly in asparagine in the white leaves (Table 3). In vitro activities of both nitrate reductase and nitrite reductase were detected in the leaf extracts of normal and chlorophyll-less leaves of albino mutant seedlings of barley (Hordeum vulgare L.), although their specific activities were less in the chlorophyll-less leaves than in the normal leaves [15].

Table 3.
Assimilation of 1 mM 15NO2− by 60-min incubation to amino acids in the green and white leaf segments from 15-day-old albino mutant rice plants in darkness.
3. Enzymes for Nitrate Assimilation for Nitrate Assimilation in Leaves
3.1. Nitrate Reduction to Ammonia
Nitrate is not only an essential nutrient but also a signaling molecule of cellular events in response to its fluctuating availability in both space and time. Nitrate is taken up into the roots by the nitrate transporters on the plasma membrane and regulates lateral root developments [16]. Transported in the xylem, nitrate is distributed within the plant by the nitrate transporters located in the shoots, leaves, flowers and seeds, and triggers expression of nitrate-responding genes, leaf development, and seed germination [17,18,19]. Nitrate reductase in the cytosol (Equation (1)) and ferredoxin (Fd)-dependent nitrite reductase in the chloroplasts/plastids (Equation (2)) catalyze the sequential reactions of nitrate reduction to nitrite and nitrite to ammonia, respectively (Figure 1).
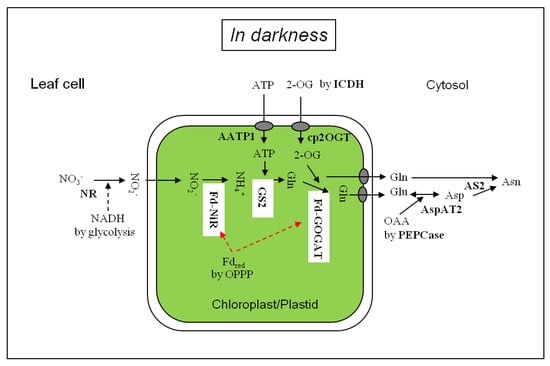
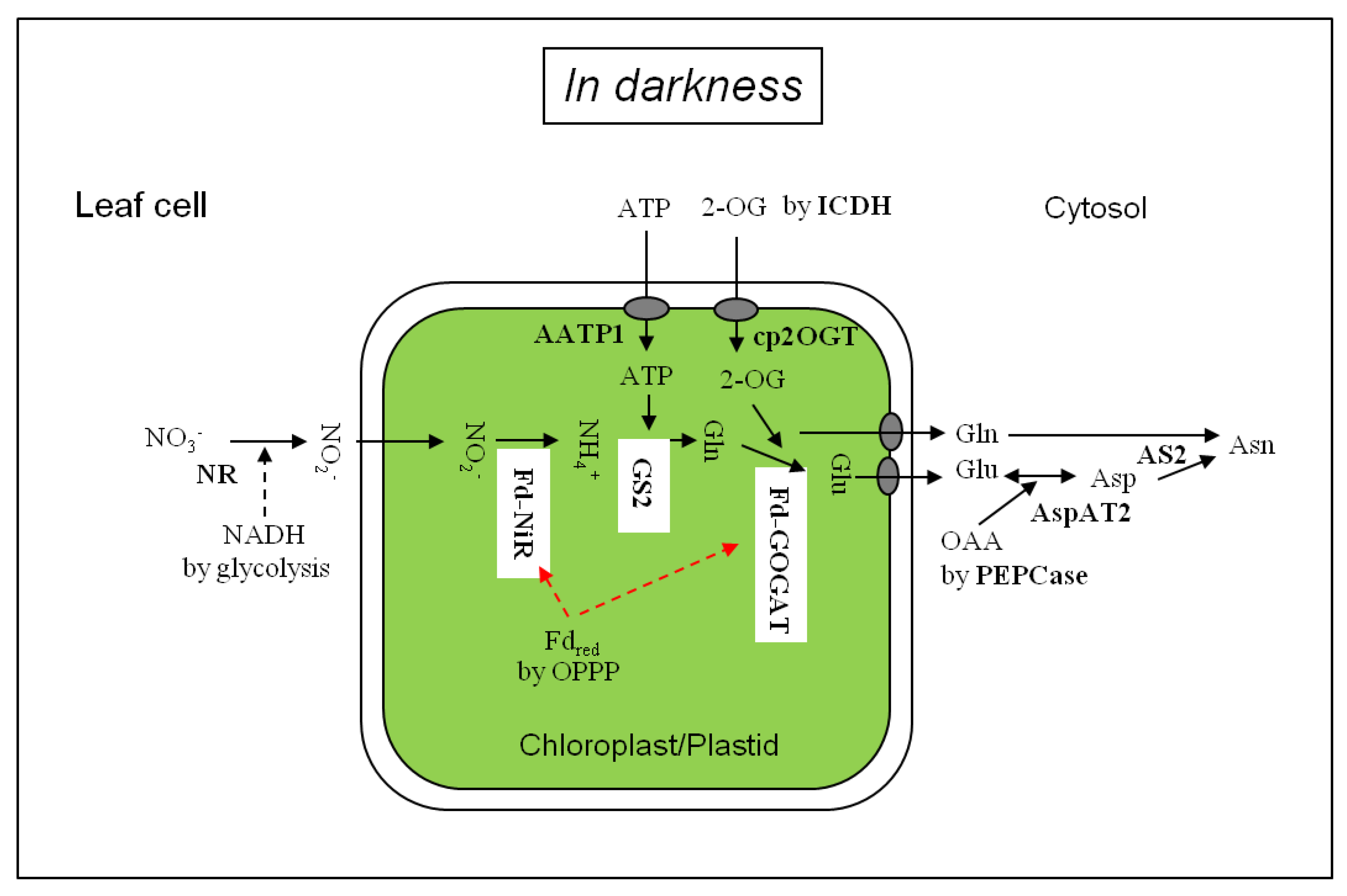
Figure 1.
Nitrate and nitrite assimilation in the leaf cells in darkness. AATP1, ATP/ADP transporter; AS, asparagine synthetase; AspAT: aspartate aminotransferase; cp2OGT, chloroplastic 2-OG transporter; Fdred, reduced ferredoxin; Fd-GOGAT, Ferredoxin-dependent glutamate synthase; Fd-NiR, Ferredoxin-dependent nitrite reductase, GS2: chloroplastic glutamine synthetase; ICDH, NADPH-isocitrate dehydrogenase; NR: nitrate reductase; PEPCase, phosphoenolpyruvate carboxylase; OAA, oxaloacetate; OPPP, oxidative pentose phosphate pathway.
Nitrate reductase (NADH-NR, EC 1.6.6.1; NAD(P)H-NR, EC 1.6.6.2)
2e
NO3− + NADH or NAD(P)H + H+ → NO2− + NAD+ or NADP+ + H2O
Nitrite reductase (NiR, EC 1.7.7.1)
6e
NO2− + 6 Fdred + 8 H+ → NH4+ + 6 Fdox + 2 H2O
NADH produced by glycolysis served as a major electron donor to nitrate reductase (NADH-NR, EC 1.6.6.1) in the leaves of most plant species [20,21], while a bi-specific nitrate reductase to NADH and NADPH (NAD(P)H-NR, EC 1.6.6.2) was found in the leaves of soybean (Glycine max. Merr.) [22,23] and barley [24]. NR, located in the cytosol as homodimer or homotetramer [25], utilizes two electrons from NAD(P)H to reduce nitrate. The enzyme has three functional domains for binding of the prosthetic group or cofactor: flavin adenine dinucleotide (FAD), heme-Fe, and molybdate-pterin (MoCo) [17,26].
Ferredoxin-dependent NiR (Fd-NiR), localized in the chloroplasts/plastids [25,27,28,29], utilizes six electrons from photo-reduced ferredoxin (Fd) as the electron donor to reduce nitrite to ammonia. Fd-NiR has two prosthetic groups: a siroheme and an iron-sulfur cluster, linked by one of four cysteine residues of the iron-sulfur cluster [26,30]. The high NiR activity, assayed by the disappearance of nitrite or formation of ammonia, was detected in the presence of strong reducing dyes methyl viologen or benzyl viologen reduced chemically by dithionite [31,32]. NiR activity in vitro was found in the presence of Fd reduced by ferredoxin: NADP+ oxidoreductase (FNR, EC 1.18.1.2) depending on NADPH, which was generated by a diaphorase containing glucose 6-phosphate dehydrogenase (G6PDH, EC 1.1.1.49) [33]. The in vitro titration analysis showed that the NiR may make a complex close to 1:1 with reduced Fd for the efficient electron transfer [34].
3.2. Glutamine Synthesis and Metabolism to Glutamate and Asparagine in Leaves
Up to 1974, it was generally accepted that ammonia assimilation is catalyzed by ammonia-inducible glutamate dehydrogenase (GDH, EC 1.4.1.2), which catalyzes a reversible amination of 2-oxoglutarate by ammonia generating L-glutamate and its conversion to ammonia and 2-oxoglutarate. Under conditions of ammonia excess, ammonia was assimilated into glutamine by glutamine synthetase (GS or L-glutamate:ammonia ligase (ADP), EC 6.3.1.2, Equation (3)), and under more excessive levels of ammonia and glutamine, asparagine served as an storage compound of nitrogen via the catalysis by glutamine-dependent asparagine synthetase (AS, EC 6.3.5.4).
However, Tempest et al. [35] reported a new pathway of ammonium assimilation by the coupled reactions of GS and NADPH-dependent glutamate synthase (L-glutamine (amide): 2-oxoglutarate aminotransferase: NADPH-GOGAT, EC 1.4.1.13) in bacteria. In plants, glutamate synthase activity with ferredoxin (Fd-GOGAT, EC 1.4.7.1) (Equation (4)) in the chloroplasts was reported by Lea and Miflin [36]. Extensive studies provided evidence for the operation of GS (GS2)/GOGAT (Fd-GOGAT) cycle in the chloroplasts/plastids as the major route of primary nitrogen assimilation [37] and photorespiratory ammonium re-assimilation in leaves [9,38,39,40,41,42].
Glutamine synthetase (GS, EC 6.3.1.2)
L-Glutamate + NH3 + ATP → L-Glutamine + ADP + Pi
Fd-glutamate synthase (Fd-GOGAT, EC 1.4.7.1)
2e
L-Glutamine + 2-Oxoglutarate + Fdred → 2 L-Glutamate + Fdox
15N tracing studies confirmed an in vivo operation of GS2/Fd-GOGAT cycle for the nitrogen assimilation from nitrate, nitrite, and ammonia into amino acids in the light or in darkness (see Table 1).
The GS occurs in two forms, cytosolic GS1 and plastidial GS2, in both leaves and roots with different ratio according to plants [43]. The cytosolic GS1 in the senescent leaves may function to assimilate a high level of ammonia during nitrogen remobilization [44,45]. Two forms of GOGAT, Fd-GOGAT and NADH-GOGAT (EC 1.4.1.14), are distinguished in leaves and roots of different plant species. The Fd-GOGAT in vitro activity was found active in the light-grown mature leaves, and the isolated chloroplasts showed a high activity of Fd-GOGAT in the light and low activity in darkness [46,47]. The enhancement of Fd-GOGAT activity and the Fd-GOGAT protein level during the greening of the etiolated plants [48] via a reversible red/far-red reaction provided evidence for a regulation mediated by the phytochromes [39,49]. Two molecules of glutamate are formed from glutamine and 2-OG through the intramolecular reactions of NH2-releasing glutaminase and 2-OG transamidation with -NH2 using reduced Fd (Equation (4)) [50,51,52].
In the cytosol of leaf cells, the amide-N of glutamine is transferred to aspartic acid to form asparagine by asparagine synthetase utilizing ATP (AS, Equation (5)). AS could use both ammonia and glutamine-amide while glutamine is a preferred amide donor. Km for glutamine (0.04–1.0 mM) was 40-fold lower than for ammonium ion [53]. The accumulation of asparagine [54] and AS-mRNA of Class I ASN genes [55,56,57] was enhanced in darkness. This is consistent with a carbon supply by an anaplerotic reaction of cytosolic phosphoenolpyruvate carboxylase [58,59]. Oxaloacetate thus formed is transaminated with glutamate by aspartate aminotransferase to aspartate, substrate of AS (AspAT, Equation (6)).
Asparagine synthetase (AS, EC 6.3.5.4)
L-Glutamine + L-Aspartate + ATP → L-Asparagine + L-Glutamate + AMP + PPi
Aspartate aminotransferase (AspAT, EC 2.6.1.1)
L-Glutamate + Oxaloacetic acid ⟷ L-Aspartate + 2-Oxoglutarate
AspAT in plants exists as isoforms, which are located in different subcellular compartments. The ASP2 mRNA for cytosolic AspAT2 in Arabidopsis was most abundantly expressed in root tissue and accumulated at higher levels in illuminated leaves and dark-adapted leaves [60], indicating that AspAT2 may be involved in synthesizing aspartate pool for asparagine synthesis by AS2 in dark-adapted plants.
Figure 1 depicts the scheme of nitrate assimilation in darkness in the leaf cells. Nitrate delivered from the xylem is reduced to NO2− by the cytosolic NR using NADH from glycolysis. Nitrite is diffused into the chloroplasts [61] and reduced to ammonia by Fd-NiR. The ammonium is assimilated to glutamine by chloroplast-localized GS2 using ATP imported [62] and then to glutamate by Fd-GOGAT using 2-OG produced via cytosolic NADP-dependent isocitrate dehydrogenase (NADPH-ICDH, EC 1.1.1.42) [63,64,65] and/or in part by mitochondrial NAD-dependent isocitrate dehydrogenase (NADH-IDH, EC 1.1.1.41) [66]. Glutamate and glutamine in the chloroplasts are released to the cytosol and glutamate is metabolized to aspartate by cytosolic AspAT2. Aspartate thus produced is combined with the amide of glutamine, forming asparagine by leaf cytosolic AS2. The nitrogen assimilation pathway from nitrate to asparagine catalyzed by Fd-NiR, GS, Fd-GOGAT, AspAT and AS in darkness in the green leaves was in line with 15NO2− tracing data shown in Table 3. In chlorophyll-less white leaves, which contained proplastids [14], the activity of AS2 (asparagine formation) might be higher than the Fd-GOGAT activity (glutamate formation), suggesting a low level of Fd-GOGAT without light [48].
4. Reductant-Supplying Systems for Dark Nitrogen Assimilation in Leaves
4.1. Reductant Supply for Nitrate Reductase and Nitrite Reductase
NADH-NR (Equation (1)) from spinach (Spinacia oleracea) had a midpoint redox potentials (Em7) of −60 mV and Em0 of NADH being −320 mV [26]. NADH can be generated in the cytosol by the respiratory network through glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EC 1.2.1.12) or through an anaplerotic reaction by malate dehydrogenase (NAD-MDH, EC 1.1.1.37) [21].
Following nitrite import from the cytosol into the chloroplasts/plastids by passive diffusion, nitrite is reduced to ammonia by NiR. This reaction involves a flow of six electrons from Fdred → ((4Fe‒4S) → siroheme) → NO2− (Equation (2)). Ferredoxins are reduced by photosystems and discovered in 1963 in the leaves of Cucurbita pepo [27] and spinach leaves [28,29]. The Em of Fd siroheme for NO2−-binding and that of (4Fe-4S) cluster of spinach NiR were determined to be −290 mV and −365 mV, respectively [67]. In maize, four Fd iso-proteins were identified showing the tissue-preferential distribution: leaf-specific and light-inducible FdI (Em = −423 mV) and FdII (Em = −406 mV) in leaves; FdIII (Em = −345 mV) and FdIV in all plant parts including roots [68,69,70,71,72] (Table 4).

Table 4.
Representative iso-proteins of Fd and FNR identified in the leaves and roots of maize and Arabidopsis.
Under the light, leaf NiR received electrons from Fd reduced in the photosystem I (PSI), while in darkness without the light energy, stromal Fd received electrons via FNR from the primary electron donor NADPH (Em = −320 mV, [76]), as shown in Figure 2. In darkness, plastidial NADPH was generated by oxidative pentose phosphate pathway using G6PDH (EC 1.1.1.49) and 6-phosphogluconate dehydrogenase (6PGDH, EC 1.1.1.44) at the expense of glucose 6-phoshate (G6P) produced from starch in the plastids and G6P imported from the cytosol to the plastids [77,78,79,80,81]. Thus, FNR (EC 1.18.1.2, Em = ~ −320 mV, [82]) in leaves and roots could catalyze the reversible electron transfer between NADPH and leaf-type Fd (e.g., FdI in maize) and root-type Fd (e.g., FdIII in maize) (see Table 4) as shown in Equation (7) [83,84]. In the leaves, both leaf-type and root-type FNRs were found in the stroma of the leaf chloroplasts as well as some leaf-type FNRs in the thylakoid membrane [73,74,75], and gene expression for the root-type FNRs was induced by nitrate feeding [18,73]. In vitro electron donation from NADPH to maize FdIII:R-FNR complex was active with a ratio of 1.00, in contrast to a lower ratio (0.68) in the leaf NADPH‒FdI:L-FNR system [85].
2Fd (Fe3+) + NADPH ⟷ 2Fd (Fe2+) + NADP+ + H+
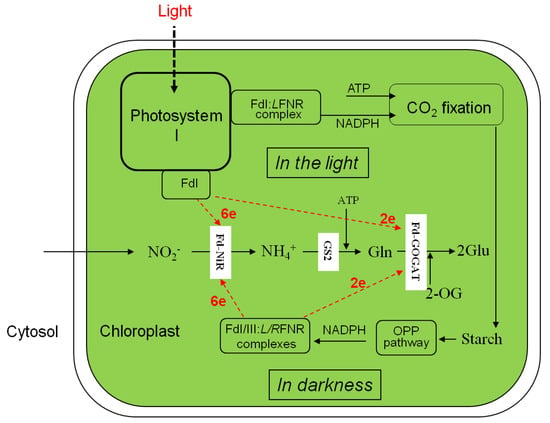
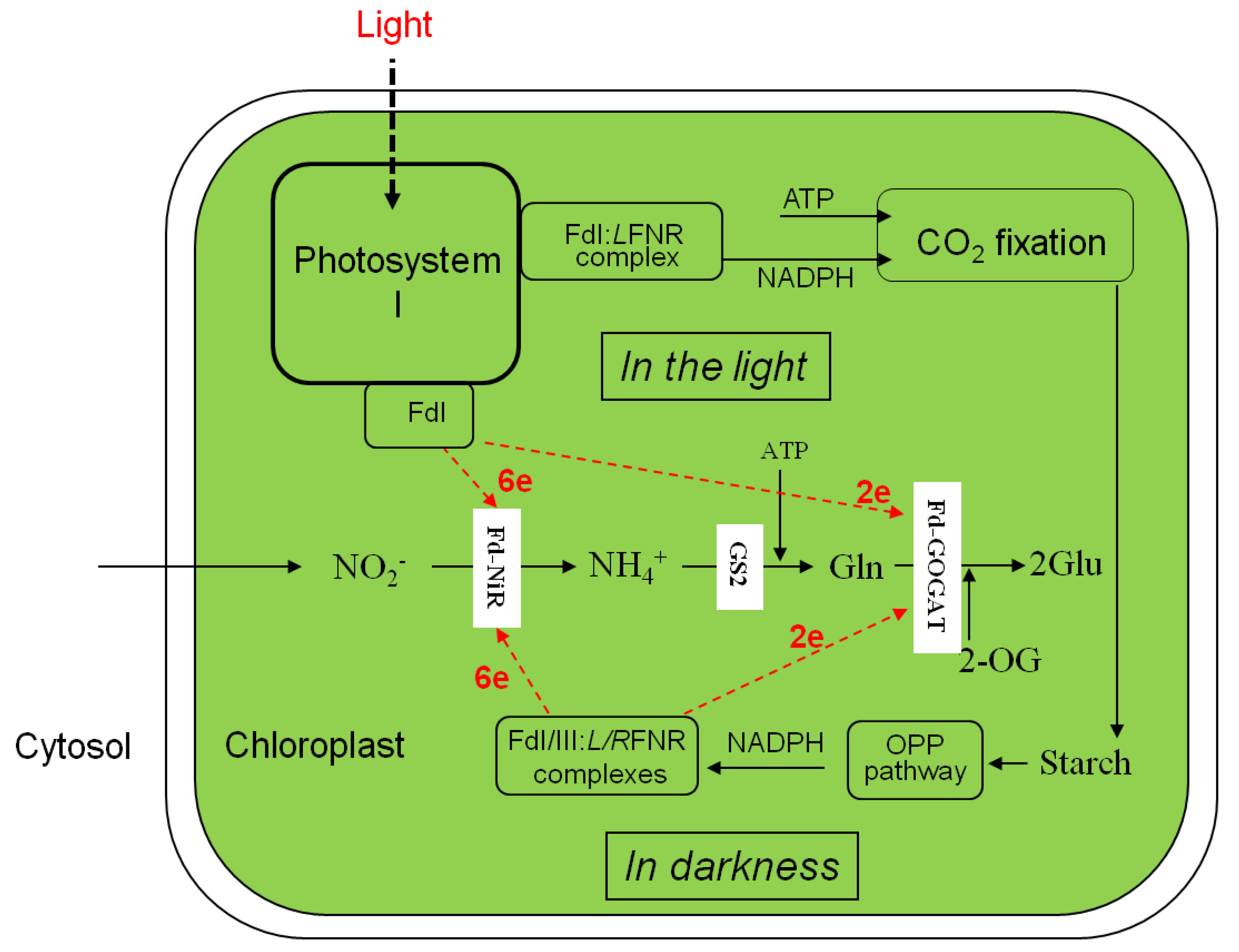
Figure 2.
Nitrite assimilation and glutamate formation in the chloroplasts in the light and darkness. LFNR, leaf-type FNR; RFNR, root-type FNR. The other abbreviations in Figure 1 legend.
The previous investigations of 15NO3− and 15NO2− reduction and assimilation into amino acids in leaf segments in darkness showed high magnitudes than in the light in tobacco [3], tomato [4], sunflower [8], and rice [14], while negligible in wheat and maize leaves [5]. Such difference in magnitudes may be derived from an availability of electron in NADPH-FNR-Fd systems under changing electron donating system in darkness (Figure 2).
Under anaerobic condition, the nitrite accumulation was large and glutamine quantity became small (Table 2). These results were caused by the elevated level of NO3−-reducing NADH and the shortage of NADPH [12,86]: The NADPH deficiency may reduce nitrite-to-ammonia reduction and induce the glutaminase activity of GOGAT, causing disappearance of glutamine [85]. Expression studies determined the level of Fd and FNR in the plants deprived of light by continuous or prolonged darkness. This dark stress declined the photosynthetic FNR subforms (LFNRI and LFNRII) at both mRNA and protein levels at the base section of wheat leaf in the presence of nitrate [87], and leaf-type Fds (Fd I and Fd II) in maize leaves [68], suggesting a less efficient contribution of NADPH-FNR-Fd system necessary to the reactions of Fd-NiR and Fd-GOGAT. Exposure of Arabidopsis to environmental stress such as extended high light (120 h at approximately 500 micromole photons m2 sec1) resulted in a gradual decrease of AtFd2 (At1g60950) in its mRNA (to 10% of the WT level) and protein (to 13%) [88]. Down-regulation or mutation of Fd in Arabidopsis [89] and potato (Solanum tuberosum) [90] caused an inactivated photosynthesis and inhibited plant growth.
4.2. Reductant Supply to the Fd-Dependent GOGAT
Ammonia produced by NiR in the plastids is assimilated into glutamine by GS2 using energy (ATP) from mitochondria [62,79]. The glutamine amide-N is transferred to 2-OG, yielding two molecules of glutamate takes place in the chloroplasts/plastids by Fd-GOGAT [36,52,91]. Fd-GOGAT is a flavin and iron-sulfur-containing protein. The isopotential of these chromophore and cluster were reported to have Em of −225 ± 10 mV in the enzyme from spinach leaves [92]. NADPH, generated by the oxidative pentose phosphate pathway, was also shown to be a primary electron donor for the reactions of Fd-GOGAT in darkness [80].
A 15N-tracing study in sunflower leaf discs (Table 1) showed that the activity of glutamine amide-N transfer to 2-oxoglutamate forming glutamate in darkness was less active than in the light, where PSI supplied electrons to Fd (see Figure 2). The activity of Fd-GOGAT in vegetable leaves was the major regulating step of nitrate assimilation in the whole plant [1].
5. Conclusions
Leaves represent a major site of primary nitrogen assimilation in concert with roots, photorespiratory NH4+ re-fixation, and translocation of nitrogen within the plant. 15N-tracing studies with leaves demonstrated that nitrate was reduced to ammonia and assimilated into glutamine, glutamate, aspartate, and asparagine in the light and in darkness. In the present review, we examined that the reductive incorporation of nitrate into amino acids occurs in darkness in the leaves through the isoforms of NR, NiR, GS, Fd-GOGAT, AspAT, and AS. To provide reducing equivalents to the NiR and Fd-GOGAT reactions in the dark, single leaf contains the photosynthetic form of FNR and Fd and heterotrophic form of FNR and Fd, indicating inter-connected electron supply systems in the light and in darkness. It remains to dissect the operation mechanism of electron donation systems in distinct types of photosynthetic cells and heterotrophic cells of a leaf.
Author Contributions
Conceptualization, writing of the original draft, reviewing and editing: T.Y. and A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank E. J. Hewitt, P. Gadal, and R. H. Hageman for their stimulating discussions for the dark nitrogen assimilation in the leaves.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yoneyama, T.; Yamada, H.; Yamagata, M.; Kouchi, H. Nitrate reduction and partitioning of nitrogen in komatsuna (Brassica campestris L. var. rapa) plants: Compartmental analysis in combination with 15N tracer experiments. Plant Cell Physiol. 1987, 28, 679–696. [Google Scholar]
- Yoneyama, T.; Suzuki, A. Exploration of nitrate-to-glutamate assimilation in non-photosynthetic roots of higher plants by studies of 15N-tracing, enzyme involved, reductant supply, and nitrate signaling: A review and synthesis. Plant Physiol. Biochem. 2019, 136, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Delwiche, C.C. The assimilation of ammonium and nitrate ions by tobacco plants. J. Biol. Chem. 1951, 189, 167–175. [Google Scholar] [PubMed]
- Mendel, J.L.; Visser, D.W. Studies on nitrate reduction in higher plants. I. Arch. Biochem. Biophys. 1951, 32, 158–169. [Google Scholar] [CrossRef]
- Canvin, D.T.; Atkins, C.A. Nitrate, nitrite and ammonia assimilation by leaves: Effect of light, carbon dioxide and oxygen. Planta 1974, 116, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Kumazawa, K. A kinetic study of the assimilation of 15N-labelled ammonium in rice seedling roots. Plant Cell Physiol. 1974, 15, 655–661. [Google Scholar] [CrossRef]
- Yoneyama, T.; Kumazawa, K. A kinetic study of the assimilation of 15N-labelled nitrate in rice seedlings. Plant Cell Physiol. 1975, 16, 21–26. [Google Scholar] [CrossRef]
- Ito, O.; Kumazawa, K. Amino acid metabolism in plant leaf. III. The effect of light on the exchange of 15N-labeled nitrogen among several amino acids in sunflower leaf discs. Soil Sci. Plant Nutr. 1978, 24, 327–336. [Google Scholar] [CrossRef]
- Keys, A.J.; Bird, I.F.; Cornelius, M.J.; Lea, P.J.; Wallsgrove, R.M.; Miflin, B.J. Photorespiratory nitrogen cycle. Nature 1978, 275, 741–743. [Google Scholar] [CrossRef]
- Klepper, L.; Flesher, D.; Hageman, R.H. Generation of reduced nicotinamide adenine dinucleotide for nitrate reductase in green leaves. Plant Physiol. 1971, 48, 580–590. [Google Scholar] [CrossRef]
- Radin, J.W. In vivo assay of nitrate reductase in cotton leaf discs. Plant Physiol. 1973, 51, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.F.; Hucklesby, D.P.; Hewitt, E.J. Effect of aerobic and anaerobic conditions on the in vitro nitrate reductase assay in spinach leaves. Planta 1979, 146, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T. 15N studies on the in vivo assay of nitrate reductase in leaves: Occurrence of underestimation of the activity due to dark assimilation of nitrate and nitrite. Plant Cell Physiol. 1981, 22, 1507–1520. [Google Scholar] [CrossRef]
- Yoneyama, T. 15N study on the dark assimilatory reduction of nitrate and nitrite in the leaf sections of chlorophyll mutants of rice (Oryza sativa L.). Plant Sci. Lett. 1984, 33, 195–200. [Google Scholar] [CrossRef]
- Sawhney, S.K.; Prakash, V.; Naik, M.S. Nitrate and nitrite reductase activities in induced chlorophyll mutants of barley. FEBS Lett. 1972, 22, 200–202. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 2000, 51, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M.; Smith, M.; Bellissimo, D.; Davis, R.W. Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc. Natl. Acad. Sci. USA 1988, 85, 5006–5010. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Evans, H.J.; Nason, A. Pyridine nucleotide-nitrate reductase from extracts of higher plants. Plant Physiol. 1953, 28, 233–254. [Google Scholar] [CrossRef]
- Emes, M.J.; Fowler, M.W. The intracellular location of the enzymes of the nitrate assimilation in the apices of seedling pea roots. Planta 1979, 144, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Jolly, S.O.; Campbell, W.; Tolbert, N.E. NADPH- and NADH-nitrate reductase from soybean leaves. Arch. Biochem. Biophys. 1976, 174, 431–439. [Google Scholar] [CrossRef]
- Orihuel-Iranzo, B.; Campbell, W.H. Development of NAD(P)H and NADH: Nitrate reductase activities in soybean cotyledons. Plant Physiol. 1980, 65, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Harker, A.R.; Narayanan, K.R.; Warner, R.L.; Kleinhofs, A. NAD(P)H bispecific nitrate reductase in barley leaves: Partial purification and characterization. Photochemistry 1986, 25, 1276–1279. [Google Scholar] [CrossRef]
- Dalling, M.J.; Tolbert, N.E.; Hageman, R.H. Intracellular location of nitrate reductase and nitrite reductase. I. Spinach and tobacco. Biochim. Biophys. Acta 1972, 283, 505–512. [Google Scholar] [CrossRef]
- Guerrero, M.G.; Vega, J.M.; Losada, M. The assimilatory nitrate-reducing system and its regulation. Ann. Rev. Plant Physiol. 1981, 32, 169–204. [Google Scholar] [CrossRef]
- Hewitt, E.J.; Betts, G.F. The reduction of nitrite and hydroxylamine by ferredoxin and chloroplast grana from Cucurbita pepo. Biochem. J. 1963, 89, 20. [Google Scholar]
- Losada, M.; Paneque, A.; Ramirez, J.M.; del Campo, F.F. Mechanism of nitrite reduction in chloroplasts. Biochem. Biophys. Res. Commun. 1963, 10, 298–303. [Google Scholar] [CrossRef]
- Huzisige, H.; Satoh, K.; Tanaka, K.; Hayasida, T. Photosynthetic nitrite reductase II. Further purification and biochemical properties of the enzyme. Plant Cell Physiol. 1963, 4, 307–322. [Google Scholar]
- Kherraz, K.; Kherraz, K.; Kameli, A. Homology modeling of ferredoxin-nitrite reductase from Arabidopsis thaliana. Bioinformation 2011, 6, 115–118. [Google Scholar] [CrossRef]
- Sanderson, G.W.; Cocking, E.C. Enzymic assimilation of nitrate in tomato plants. II. Reduction of nitrite to ammonia. Plant Physiol. 1964, 39, 423–431. [Google Scholar] [CrossRef]
- Cresswell, C.F.; Hageman, R.H.; Hewitt, E.J.; Hucklesby, D.P. The reduction of nitrate, nitrite and hydroxylamine to ammonia by enzymes from Cucurbita pepo L. in the presence of reduced benzyl viologen as electron donor. Biochem. J. 1965, 94, 40–53. [Google Scholar] [CrossRef]
- Joy, K.W.; Hageman, R.H. The purification and properties of nitrite reductase from higher plants, and its dependence on ferredoxin. Biochem. J. 1966, 100, 263–273. [Google Scholar] [CrossRef]
- Knaff, D.B.; Smith, J.M.; Malkin, R. Complex formation between ferredoxin and nitrite reductase. FEBS Lett. 1978, 90, 195–197. [Google Scholar] [CrossRef]
- Tempest, D.W.; Meers, J.L.; Brown, C.M. Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem. J. 1970, 117, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.J.; Miflin, B.J. Alternative route for nitrogen assimilation in higher plants. Nature 1974, 251, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Potel, F.; Valadier, M.-H.; Ferrario-Méry, S.; Grandjean, O.; Morin, H.; Gaufichon, L.; Boutet-Mercey, S.; Lothier, J.; Rothstein, S.J.; Hirose, N.; et al. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana—Roles of glutamate synthases and carbamoylphosphate synthase in leaves. FEBS J. 2009, 276, 4061–4076. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.R.; Ogren, W.L. Inhibition of photosynthesis in Arabidopsis mutants lacking in leaf glutamate synthase activity. Nature 1980, 286, 257–259. [Google Scholar] [CrossRef]
- Ziegler, C.; Feraud, M.; Jouglet, T.; Viret, L.; Spampinato, A.; Paganelli, V.; Ben Hammouda, M.; Suzuki, A. Regulation of promoter activity of ferredoxin-dependent glutamate synthase. Plant Physiol. Biochem. 2003, 41, 649–655. [Google Scholar] [CrossRef]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes: Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 2003, 41, 555–564. [Google Scholar] [CrossRef]
- Hirel, B.; Krapp, A. Nitrogen utilization in plants I Biological and agronomic importance. In Encyclopedia of Biochemistry, 3rd ed.; Lennarz, W., Lane, M., Eds.; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Hirel, B.; Gadal, P. Glutamine synthetase in rice. A comparative study of the enzymes from roots and leaves. Plant Physiol. 1980, 66, 619–623. [Google Scholar] [CrossRef]
- Lothier, J.; Gaufichon, L.; Sormani, R.; Lemaître, T.; Azzopardi, M.; Morin, H.; Chardon, F.; Reisdorf-Cre, M.; Avice, J.-C.; Masclaux-Daubresse, C. The cytosolic glutamine synthetase GLN1; 2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J. Exp. Bot. 2011, 62, 1375–1390. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; de Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic glutamine synthase Gln1; 2 is the main isozyme contributing to GS1 activity and can be up-regulated to relieve ammonium toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef]
- Matoh, T.; Takahashi, E. Changes in the activities of ferredoxin- and NADH-glutamate synthase during seedling development of peas. Planta 1982, 154, 289–294. [Google Scholar] [CrossRef]
- Wallsgrove, R.M.; Lea, P.J.; Miflin, B.J. The development of NAD(P)H-dependent and ferredoxin-dependent glutamate synthase in greening barley and pea leaves. Planta 1982, 154, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Rothstein, S. Structure and regulation of ferredoxin-dependent glutamate synthase from Arabidopsis thaliana. Cloning of cDNA, expression in different tissues of wild-type and gltS mutant strains, and light induction. Eur. J. Biochem. 1997, 243, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Rioual, S.; Lemarchand, S.; Godfroy, N.; Roux, Y.; Boutin, J.-P.; Rothstein, S. Regulation by light and metabolites of ferredoxin-dependent glutamate synthase in maize. Physiol. Plant. 2001, 112, 524–530. [Google Scholar] [CrossRef]
- Suzuki, A.; Vidal, J.; Gadal, P. Glutamate synthase isoforms in rice. Immunological studies of enzymes in green leaf, etiolated leaf, and root tissues. Plant Physiol. 1982, 70, 827–832. [Google Scholar] [CrossRef]
- Suzuki, A.; Gadal, P. Glutamate synthase: Physicochemical and functional properties of different forms in higher plants and in other organisms. Physiol. Vég. 1984, 22, 471–486. [Google Scholar]
- Suzuki, A.; Knaff, D.G. Glutamate synthase: Structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth. Res. 2005, 83, 191–217. [Google Scholar] [CrossRef]
- Huber, T.A.; Streeter, J.G. Asparagine biosynthesis in soybean nodules. Plant Physiol. 1984, 74, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Joy, K.W.; Ireland, R.J.; Lea, P.J. Asparagine synthesis in pea leaves, and the occurrence of an asparagine synthetase inhibitor. Plant Physiol. 1983, 73, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.-Y.; Coruzzi, G. Dark-induced and organ-specific expression of two asparagine synthetase genes in Pisum sativum. EMBO J. 1990, 9, 323–332. [Google Scholar] [CrossRef]
- Tsai, F.-Y.; Coruzzi, G. Light represses transcription of asparagine synthetase genes in photosynthetic and non-photosynthetic organs. Mol. Cell. Biol. 1991, 11, 4966–4972. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine metabolic pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef]
- Kirk, P.R.; Leech, R.M. Amino acid biosynthesis by isolated chloroplasts during photosynthesis. Plant Physiol. 1972, 50, 228–234. [Google Scholar] [CrossRef]
- Melzer, E.; O’Leary, M.H. Anaplerotic CO2 fixation by phosphoenolpyruvate carboxylase in C3 plants. Plant Physiol. 1987, 84, 58–60. [Google Scholar] [CrossRef]
- Schultz, C.J.; Hsu, M.; Hiesak, B.; Coruzzi, G.M. Arabidopsis mutants define an in Vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 1998, 149, 491–499. [Google Scholar]
- Shingles, R.; Roh, M.H.; McCarty, R.E. Nitrite transport in chloroplast inner envelop vesicles. Plant Physiol. 1996, 112, 1375–1381. [Google Scholar] [CrossRef][Green Version]
- Schünemann, D.; Borchert, S.; Flügge, U.-I.; Heldt, H.W. ADP/ATP translocator from pea root plastids. Comparison with translocation from spinach chloroplasts and pea leaf mitochondria. Plant Physiol. 1993, 103, 131–137. [Google Scholar] [CrossRef]
- Kruse, A.; Fieuw, S.; Heineke, D.; Müller-Röber, B. Antisense inhibition of cytosolic NADP-dependent isocitrate dehydrogenase in transgenic potato plants. Planta 1998, 205, 82–91. [Google Scholar] [CrossRef]
- Lancien, M.; Gadal, P.; Hodges, M. Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol. 2000, 123, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Sulpice, R.; Sienkiewicz-Porzucek, A.; Osorio, S.; Krahnert, I.; Stitt, M.; Fernie, A.R.; Nues-Nesi, A. Mild reduction in cytosolic NADP-dependent isocitrate dehydrogenase activity in lower amino acid contents and pigmentation without impacting growth. Amino Acids 2010, 39, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-D.; Gadal, P. Do mitochondria provide the 2-oxoglutarate needed for glutamate synthesis in higher plant chloroplasts? Plant Physiol. Biochem. 1990, 28, 141–145. [Google Scholar]
- Hirasawa, M.; Tollin, G.; Salamon, Z.; Knaff, D.B. Transient kinetic and oxidation-reduction studies of spinach ferredoxin:nitrite oxidoreductase. Biochim. Biophys. Acta 1994, 1185, 336–345. [Google Scholar] [CrossRef]
- Kimata, Y.; Hase, T. Localization of ferredoxin isoproteins in mesophyll and bundle sheath cells in maize leaf. Plant Physiol. 1989, 89, 1193–1197. [Google Scholar] [CrossRef]
- Matsumura, T.; Kimata-Ariga, Y.; Sakakibara, H.; Sugiyama, T.; Murata, H.; Takao, T.; Shimonishi, Y.; Hase, T. Complementary DNA cloning and characterization of ferredoxin localized in bundle-sheath cells of maize leaves. Plant Physiol. 1999, 119, 481–488. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Onda, Y.; Ashikari, T.; Tanaka, Y.; Kusumi, T.; Hase, T. Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 2000, 122, 887–894. [Google Scholar] [CrossRef]
- Onda, Y.; Matsumura, T.; Kimata-Ariga, Y.; Sakakibara, H.; Sugiyama, T.; Hase, T. Differential interaction of maize root ferredoxin: NADP+ oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 2000, 123, 1037–1045. [Google Scholar] [CrossRef]
- Shinohara, F.; Kurisu, G.; Hanke, G.; Bowsher, C.; Hase, T.; Kimata-Ariga, Y. Structural basis for the isotype-specific interaction of ferredoxin and ferredoxin: NADP+ oxidoreductase: An evolutionary switch between photosynthetic and heterotrophic assimilation. Photosynth. Res. 2017, 134, 281–289. [Google Scholar] [CrossRef]
- Sakakibara, H. Differential response of genes for ferredoxin and ferredoxin: NADP+ oxidoreductase to nitrate and light in maize leaves. J. Plant Physiol. 2003, 160, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Okutani, S.; Hanke, G.T.; Satomi, Y.; Takao, T.; Kurisu, G.; Suzuki, A.; Hase, T. Three maize leaf ferredoxin: NADPH oxidoreductase vary in subchloroplast location, expression, and interaction with ferredoxin. Plant Physiol. 2005, 139, 1451–1459. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hanke, G.T.; Okutani, S.; Satomi, Y.; Takao, T.; Suzuki, A.; Hase, T. Multiple iso-proteins of FNR in Arabidopsis: Evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ. 2005, 28, 1146–1157. [Google Scholar] [CrossRef]
- Arnon, D.I. The discovery of ferredoxin: The photosynthetic path. Trends Biochem. Sci. 1988, 13, 30–33. [Google Scholar] [CrossRef]
- Miflin, B.J.; Beevers, H. Isolation of intact plastids from a range of plant tissues. Plant Physiol. 1974, 53, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Wendt, U.K.; Wenderoth, I.; Tegeler, A.; von Schaewen, A. Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J. 2000, 23, 723–733. [Google Scholar] [CrossRef]
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Ann. Rev. Plant Physiol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Bowsher, C.G.; Tobin, A.K. Compartmentation of metabolism within mitochondria and plastids. J. Exp. Bot. 2001, 52, 513–527. [Google Scholar] [CrossRef]
- Wakao, S.; Benning, C. Genome-wide analysis of glucose-6-phosphate dehydrogenase in Arabidopsis. Plant J. 2005, 41, 243–256. [Google Scholar] [CrossRef]
- Keirns, J.J.; Wang, J.H. Studies on nicotinamide adenine dinucreotide phosphate reductase of spinach chloroplasts. J. Biol. Chem. 1972, 247, 7374–7382. [Google Scholar] [PubMed]
- Arakaki, A.K.; Ceccarelli, E.A.; Carrillo, N. Plant-type ferredoxin-NADP+ reductase: A basal structural framework and multiplicity of functions. FASEB J. 1997, 11, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, N.; Ceccarelli, E.A. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 2003, 270, 1900–1915. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Fujimori, T.; Yanagisawa, S.; Hase, T.; Suzuki, A. 15N tracing studies on in vitro reactions of ferredoxin-dependent nitrite reductase and glutamate synthase using reconstituted electron donation systems. Plant Cell Physiol. 2015, 56, 1154–1161. [Google Scholar] [CrossRef]
- Watt, M.P.; Gray, V.M.; Cresswell, C.F. Control of nitrate and nitrite assimilation by carbohydrate reserves, adenosine nucleotides and pyridine nucleotides in leaves of Zea mays L. under dark conditions. Planta 1987, 172, 548–554. [Google Scholar] [CrossRef]
- Gummadova, J.O.; Fletcher, G.J.; Moolna, A.; Hanke, G.T.; Hase, T.; Bowsher, C.G. Expression of multiple forms of ferredoxin NADP+ oxidoreductase in wheat leaves. J. Exp. Bot. 2007, 58, 3971–3985. [Google Scholar] [CrossRef]
- Liu, J.; Wang, P.; Liu, B.; Feng, D.; Zhang, J.; Su, J.; Zhang, Y.; Wang, J.-F.; Wang, H.-B. A deficiency in chloroplastic ferredoxin 2 facilitates effective photosynthetic capacity during long-term high light acclimation in Arabidopsis thaliana. Plant J. 2013, 76, 861–874. [Google Scholar] [CrossRef]
- Voss, I.; Goss, T.; Murozuka, E.; Altmann, B.; McLean, K.J.; Rigby, S.E.; Munro, A.W.; Scheibe, R.; Hase, T.; Hanke, G.T. FdC1, a novel ferredoxin protein capable of alternative electron partitioning, increases in conditions of acceptor limitation at photosystem I. J. Biol. Chem. 2011, 286, 50–59. [Google Scholar] [CrossRef]
- Holtgrefe, S.; Bader, K.P.; Horton, P.; Scheibe, R.; von Schaewen, A.; Backhausen, J.E. Decreased content of leaf ferredoxin changes electron distribution and limits photosynthesis in transgenic potato plants. Plant Physiol. 2003, 133, 1768–1778. [Google Scholar] [CrossRef]
- Vanoni, M.A.; Curti, B. Glutamate synthase: A complex iron-sulfur flavoprotein. Cell Mol. Life Sci. 1999, 55, 617–638. [Google Scholar] [CrossRef]
- Hirasawa, M.; Murley, J.K.; Salamon, Z.; Tollin, G.; Knaff, D.B. Oxidation-reduction and transient kinetic studies of spinach ferredoxin-dependent glutamate synthase. Arch. Biochem. Biophys. 1996, 330, 209–215. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).