H3.1 Eviction Marks Female Germline Precursors in Arabidopsis
Abstract
:1. Introduction
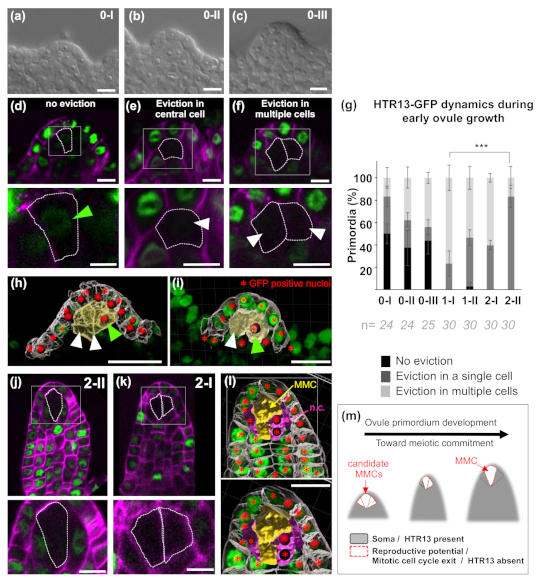
2. Results
3. Discussion
4. Material and Methods
4.1. Plant Materials
4.2. Microscopy and Image Analysis
4.3. Quantifications and Image Analysis.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Webb, M.C.; Gunning, B.E.S. Embryo sac development in Arabidopsis thaliana. Sex. Plant Reprod. 1990, 3, 244–256. [Google Scholar] [CrossRef]
- Bowman, J.L.E. Arabidopsis: An Atlas of Morphology and Development; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1993. [Google Scholar]
- Schneitz, K.; Hülskamp, M.; Pruitt, R.E. Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 1995, 7, 731–749. [Google Scholar] [CrossRef]
- Lora, J.; Yang, X.; Tucker, M.R. Establishing a framework for female germline initiation in the plant ovule. J. Exp. Bot. 2019, 70, 2937–2949. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Leon-Martinez, G.; Abad-Vivero, U.; Vielle-Calzada, J.P. Natural variation in epigenetic pathways affects the specification of female gamete precursors in arabidopsis. Plant Cell 2015, 27, 1034–1045. [Google Scholar] [CrossRef] [Green Version]
- Pinto, S.C.; Mendes, M.A.; Coimbra, S.; Tucker, M.R. Revisiting the female germline and its expanding toolbox. Trends Plant Sci. 2019, 24, 455–467. [Google Scholar] [CrossRef]
- Zhao, X.; Bramsiepe, J.; Van Durme, M.; Komaki, S.; Prusicki, M.A.; Maruyama, D.; Forner, J.; Medzihradszky, A.; Wijnker, E.; Harashima, H.; et al. Retinoblastoma related1 mediates germline entry in arabidopsis. Science 2017, 356, 6336. [Google Scholar] [CrossRef]
- Cao, L.; Wang, S.; Venglat, P.; Zhao, L.; Cheng, Y.; Ye, S.; Qin, Y.; Datla, R.; Zhou, Y.; Wang, H. Arabidopsis ick/krp cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLoS Genet. 2018, 14, e1007230. [Google Scholar] [CrossRef]
- Olmedo-Monfil, V.; Duran-Figueroa, N.; Arteaga-Vazquez, M.; Demesa-Arevalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.P. Control of female gamete formation by a small rna pathway in arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef]
- Schmidt, A.; Schmid, M.W.; Grossniklaus, U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 2015, 142, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Extavour, C.G.; Akam, M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 2003, 130, 5869–5884. [Google Scholar] [CrossRef] [Green Version]
- Kimble, J. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a002683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Western, P.S.; Miles, D.C.; van den Bergen, J.A.; Burton, M.; Sinclair, A.H. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 2008, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Wolgemuth, D.J.; Roberts, S.S. Regulating mitosis and meiosis in the male germ line: Critical functions for cyclins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1653–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, W.; Grimanelli, D.; Rutowicz, K.; Whitehead, M.W.; Puzio, M.; Kotlinski, M.; Jerzmanowski, A.; Baroux, C. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 2013, 140, 4008–4019. [Google Scholar] [CrossRef] [Green Version]
- Hernandez Lagana, E.; Mosca, G.; Mendocilla-Sato, E.; Pires, N.; Giraldo-Fonseca, A.; Grossniklaus, U.; Hamant, O.; Godin, C.; Boudaoud, A.; Grimanelli, D.; et al. Developmental constraints modulate reproductive fate and plasticity within the arabidopsis ovule. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ingouff, M.; Rademacher, S.; Holec, S.; Soljić, L.; Xin, N.; Readshaw, A.; Foo, S.H.; Lahouze, B.; Sprunck, S.; Berger, F. Zygotic resetting of the histone 3 variant repertoire participates in epigenetic reprogramming in arabidopsis. Curr. Biol. 2010, 20, 2137–2143. [Google Scholar] [CrossRef] [Green Version]
- Okada, T.; Endo, M.; Singh, M.B.; Bhalla, P.L. Analysis of the histone h3 gene family in arabidopsis and identification of the male-gamete-specific variant atmgh3. Plant J. 2005, 44, 557–568. [Google Scholar] [CrossRef]
- Otero, S.; Desvoyes, B.; Peiró, R.; Gutierrez, C. Histone h3 dynamics reveal domains with distinct proliferation potential in the arabidopsis root. Plant Cell 2016, 28, 1361–1371. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.; Wuest, S.E.; Vijverberg, K.; Baroux, C.; Kleen, D.; Grossniklaus, U. Transcriptome analysis of the arabidopsis megaspore mother cell uncovers the importance of rna helicases for plant germline development. PLoS Biol. 2011, 9, e1001155. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Wang, N.; Hou, Z.; Li, B.; Li, D.; Liu, Y.; Cai, H.; Qin, Y.; Chen, X. Regulation of female germline specification via small rna mobility in arabidopsis. Plant Cell 2020, 32, 2842–2854. [Google Scholar] [CrossRef]
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendía, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of h3k27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.V.; Desvoyes, B.; Gutierrez, C. Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 2020, 71, 5191–5204. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Gafni, O.; Weinberger, L.; Zviran, A.; Ayyash, M.; Rais, Y.; Krupalnik, V.; Zerbib, M.; Amann-Zalcenstein, D.; Maza, I.; et al. The h3k27 demethylase utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012, 488, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Surani, M.A. Germline and pluripotent stem cells. Cold Spring Harb. Perspect. Biol. 2015, 7, a019422. [Google Scholar] [CrossRef] [Green Version]
- Blagosklonny, M.V.; Pardee, A.B. The restriction point of the cell cycle. Cell Cycle 2002, 1, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Neganova, I.; Lako, M. G1 to s phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008, 213, 30–44. [Google Scholar] [CrossRef]
- Tessadori, F.; van Zanten, M.; Pavlova, P.; Clifton, R.; Pontvianne, F.; Snoek, L.B.; Millenaar, F.F.; Schulkes, R.K.; van Driel, R.; Voesenek, L.A.; et al. Phytochrome b and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000638. [Google Scholar] [CrossRef] [Green Version]
- Desvoyes, B.; Fernández-Marcos, M.; Sequeira-Mendes, J.; Otero, S.; Vergara, Z.; Gutierrez, C. Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 2014, 5, 369. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Berger, F. DNA replication-coupled histone modification maintains polycomb gene silencing in plants. Science 2017, 357, 1146–1149. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Walbot, V. Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 2014, 77, 639–652. [Google Scholar] [CrossRef] [Green Version]
- Skinner, D.J.; Hill, T.A.; Gasser, C.S. Regulation of ovule development. Plant Cell 2004, 16, S32–S45. [Google Scholar] [CrossRef] [PubMed]
- Mendocilla-Sato, E.; Baroux, C. Analysis of 3d cellular organization of fixed plant tissues using a user-guided platform for image segmentation. Bio Protocol 2017, 7. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez-Lagana, E.; Autran, D. H3.1 Eviction Marks Female Germline Precursors in Arabidopsis. Plants 2020, 9, 1322. https://doi.org/10.3390/plants9101322
Hernandez-Lagana E, Autran D. H3.1 Eviction Marks Female Germline Precursors in Arabidopsis. Plants. 2020; 9(10):1322. https://doi.org/10.3390/plants9101322
Chicago/Turabian StyleHernandez-Lagana, Elvira, and Daphné Autran. 2020. "H3.1 Eviction Marks Female Germline Precursors in Arabidopsis" Plants 9, no. 10: 1322. https://doi.org/10.3390/plants9101322



