Detailed Genetic Analysis for Identifying QTLs Associated with Drought Tolerance at Seed Germination and Seedling Stages in Barley
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Variation in Germination and Seedling Traits
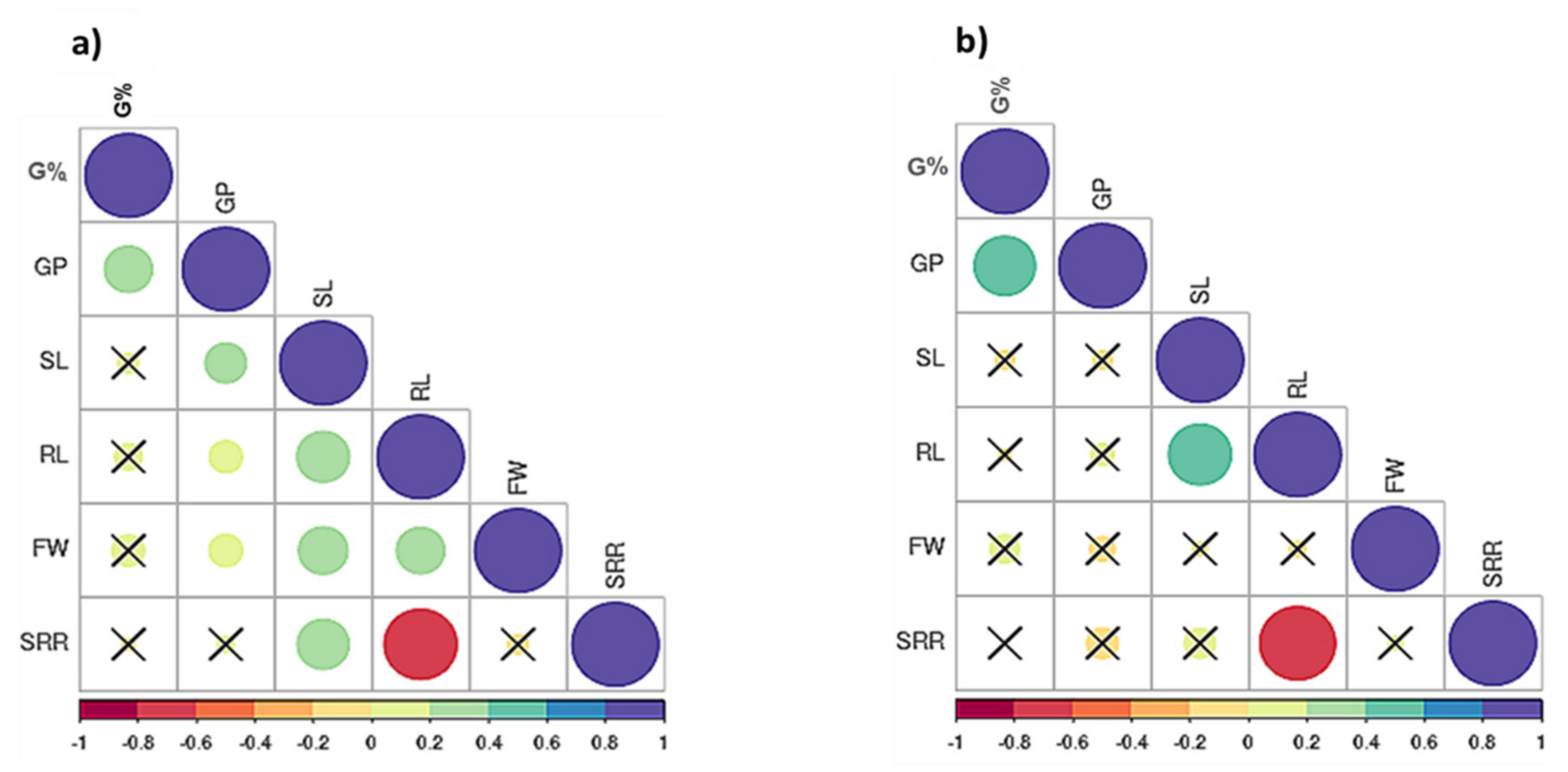
2.2. Correlation Analysis
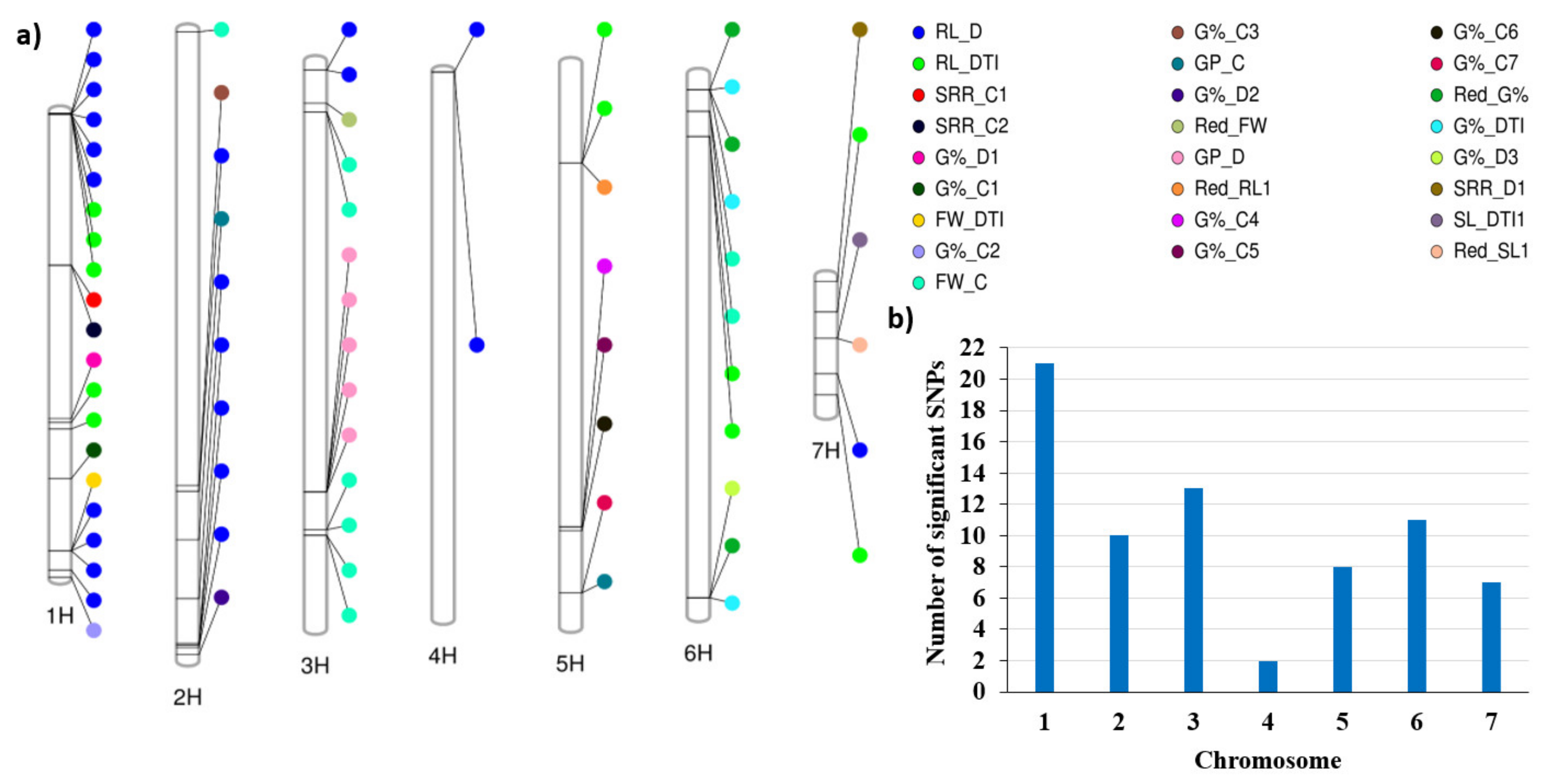
2.3. Marker–Trait Association (QTL Analysis)
2.3.1. QTLs for G% Traits
2.3.2. QTLs for GP Traits
2.3.3. QTLs for RL Traits
2.3.4. QTLs for SL Traits
2.3.5. QTLs for FW Traits
2.3.6. QTLs for SRR Traits
2.4. QTLs Associated with Drought Tolerance at Different Growth Stages
3. Discussion
3.1. Genetic Variation in Drought Tolerance at Germination and Seedling Stages
3.2. Phenotypic Correlation
3.3. The Marker–Trait Association
3.4. Marker Validation
3.5. Analysis of Gene Annotation
4. Materials and Methods
4.1. Plant Material
Seed Germination Test
4.2. Data Analyses
4.3. DNA Extraction and Genotyping-by-Sequencing (GBS)
4.4. Candidate Gene Identification
4.5. QTLs Controlling Drought Tolerance at Different Growth Stages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Nazari, L.; Pakniyat, H. Assessment of drought tolerance in barley genotypes. J. Appl. Sci. 2010, 10, 151–156. [Google Scholar] [CrossRef]
- Bourgne, S.; Job, C.; Job, D. Sugarbeet seed priming: Solubilization of the basic subunit of 11-S globulin in individual seeds. Seed Sci. Res. 2000, 10, 153–161. [Google Scholar] [CrossRef]
- Baum, M.; Von Korff, M.; Guo, P.; Lakew, B.; Hamwieh, A.; Lababidi, S.; Udupa, S.M.; Sayed, H.; Choumane, W.; Grando, S. Molecular approaches and breeding strategies for drought tolerance in barley. In Genomics-Assisted Crop Improvement; Springer: Dordrecht, The Netherlands, 2007; pp. 51–79. [Google Scholar]
- Hackenberg, M.; Gustafson, P.; Langridge, P.; Shi, B.J. Differential expression of micro RNA s and other small RNA s in barley between water and drought conditions. Plant Biotechnol. J. 2015, 13, 2–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, D. The effects of manure, genotype, seed priming, depth and date of sowing on the emergence and early growth of Sorghum bicolor (L.) Moench in semi-arid Botswana. Soil Tillage Res. 1996, 40, 73–88. [Google Scholar]
- Chen, K.; Arora, R.; Arora, U. Osmopriming of spinach (Spinacia oleracea L. cv. Bloomsdale) seeds and germination performance under temperature and water stress. Seed Sci. Technol. 2010, 38, 36–48. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Karam, M.A.; Graner, A.; Alqudah, A.M. Genetic basis of drought tolerance during seed germination in barley. PLoS ONE 2018, 13, e0206682. [Google Scholar] [CrossRef]
- Wehner, G.G.; Balko, C.C.; Enders, M.M.; Humbeck, K.K.; Ordon, F.F. Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biol. 2015, 15, 125. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Ghani, A.H.; Sharma, R.; Wabila, C.; Dhanagond, S.; Owais, S.J.; Duwayri, M.A.; Al-Dalain, S.A.; Klukas, C.; Chen, D.; Lübberstedt, T. Genome-wide association mapping in a diverse spring barley collection reveals the presence of QTL hotspots and candidate genes for root and shoot architecture traits at seedling stage. BMC Plant Biol. 2019, 19, 216. [Google Scholar] [CrossRef]
- Hittalmani, S.; Huang, N.; Courtois, B.; Venuprasad, R.; Shashidhar, H.; Zhuang, J.; Zheng, K.; Liu, G.; Wang, G.; Sidhu, J. Identification of QTL for growth-and grain yield-related traits in rice across nine locations of Asia. Theor. Appl. Genet. 2003, 107, 679–690. [Google Scholar] [CrossRef]
- Chloupek, O.; Dostál, V.; Středa, T.; Psota, V.; Dvořáčková, O. Drought tolerance of barley varieties in relation to their root system size. Plant Breed. 2010, 129, 630–636. [Google Scholar] [CrossRef]
- Reinert, S.; Kortz, A.; Léon, J.; Naz, A.A. Genome-wide association mapping in the global diversity set reveals new QTL controlling root system and related shoot variation in barley. Front. Plant Sci. 2016, 7, 1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, H.; Hickey, L.; Richard, C.; Mace, E.; Kelly, A.; Borrell, A.; Franckowiak, J.; Fox, G. Genomic regions influencing seminal root traits in barley. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Tarawneh, R.A. Mapping and identifying genes for drought tolerance in barley (Hordeum vulgare L.). Ph.D. Thesis, Der Martin-Luther-Universität Halle-Wittenberg, Halle, Germany, 2020. [Google Scholar] [CrossRef]
- Fakheri, B.A.; Aghnoum, R.; Nezhad, N.M.; Ataei, R. GWAS analysis in spring barley (Hordeum vulgare L.) for morphological traits exposed to drought. PLoS ONE 2018, 13, e0204952. [Google Scholar]
- Hansen, L.; von Wettstein-Knowles, P. The barley genes Acl1 and Acl3 encoding acyl carrier proteins I and III are located on different chromosomes. Mol. Gen. Genet. MGG 1991, 229, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Teulat, B.; Monneveux, P.; Wery, J.; Borries, C.; Souyris, I.; Charrier, A.; This, D. Relationships between relative water content and growth parameters under water stress in barley: A QTL study. New Phytol. 1997, 137, 99–107. [Google Scholar] [CrossRef]
- Teulat, B.; This, D.; Khairallah, M.; Borries, C.; Ragot, C.; Sourdille, P.; Leroy, P.; Monneveux, P.; Charrier, A. Several QTLs involved in osmotic-adjustment trait variation in barley (Hordeum vulgare L.). Theor. Appl. Genet. 1998, 96, 688–698. [Google Scholar] [CrossRef]
- Teulat, B.; Borries, C.; This, D. New QTLs identified for plant water status, water-soluble carbohydrate and osmotic adjustment in a barley population grown in a growth-chamber under two water regimes. Theor. Appl. Genet. 2001, 103, 161–170. [Google Scholar] [CrossRef]
- Sayed, M.A.; Schumann, H.; Pillen, K.; Naz, A.A.; Léon, J. AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet. 2012, 13, 61. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Shabala, S.; Ma, Y.; Xu, R.; Zhou, M. Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genom. 2015, 16, 43. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.M.; Dai, H.; Zheng, W.; Cao, F.; Zhang, G.; Sun, D.; Wu, F. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2013, 63, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Wójcik-Jagła, M.; Fiust, A.; Kościelniak, J.; Rapacz, M. Association mapping of drought tolerance-related traits in barley to complement a traditional biparental QTL mapping study. Theor. Appl. Genet. 2018, 131, 167–181. [Google Scholar] [CrossRef] [Green Version]
- Baum, M.; Grando, S.; Backes, G.; Jahoor, A.; Sabbagh, A.; Ceccarelli, S. QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’× H. spontaneum 41-1. Theor. Appl. Genet. 2003, 107, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Diab, A.A.; Teulat-Merah, B.; This, D.; Ozturk, N.Z.; Benscher, D.; Sorrells, M.E. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor. Appl. Genet. 2004, 109, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Baum, M.; Varshney, R.K.; Graner, A.; Grando, S.; Ceccarelli, S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 2008, 163, 203–214. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakew, B.; Henry, R.J.; Ceccarelli, S.; Grando, S.; Eglinton, J.; Baum, M. Genetic analysis and phenotypic associations for drought tolerance in Hordeum spontaneum introgression lines using SSR and SNP markers. Euphytica 2013, 189, 9–29. [Google Scholar] [CrossRef]
- Rollins, J.A.; Drosse, B.; Mulki, M.; Grando, S.; Baum, M.; Singh, M.; Ceccarelli, S.; von Korff, M. Variation at the vernalisation genes Vrn-H1 and Vrn-H2 determines growth and yield stability in barley (Hordeum vulgare) grown under dryland conditions in Syria. Theor. Appl. Genet. 2013, 126, 2803–2824. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Samarah, N.H.; Mullen, R.E. Drought stress effect on crop pollination, seed set, yield and quality. In Alternative Farming Systems, Biotechnology, Drought Stress and Ecological Fertilisation; Springer: Dordrecht, The Netherlands, 2011; pp. 193–213. [Google Scholar]
- Varshney, R.; Paulo, M.; Grando, S.; Van Eeuwijk, F.; Keizer, L.; Guo, P.; Ceccarelli, S.; Kilian, A.; Baum, M.; Graner, A. Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Field Crops Res. 2012, 126, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Forster, B.; Ellis, R.; Moir, J.; Talame, V.; Sanguineti, M.; Tuberosa, R.; This, D.; Teulat-Merah, B.; Ahmed, I.; Mariy, S. Genotype and phenotype associations with drought tolerance in barley tested in North Africa. Ann. Appl. Biol. 2004, 144, 157–168. [Google Scholar] [CrossRef]
- Mikolajczak, K.; Kuczynska, A.; Krajewski, P.; Sawikowska, A.; Surma, M.; Ogrodowicz, P.; Adamski, T.; Krystkowiak, K.; Gorny, A.G.; Kempa, M. Quantitative trait loci for plant height in Maresi x CamB barley population and their associations with yield-related traits under different water regimes. J. Appl. Genet. 2017, 58, 1. [Google Scholar] [CrossRef] [Green Version]
- Von Korff, M.; Grando, S.; Del Greco, A.; This, D.; Baum, M.; Ceccarelli, S. Quantitative trait loci associated with adaptation to Mediterranean dryland conditions in barley. Theor. Appl. Genet. 2008, 117, 653–669. [Google Scholar] [CrossRef]
- Boyer, J.S.; Kramer, P.J. Water Relations of Plants and Soils; Academic Press, Inc.: Cambridge, MA, USA, 1995. [Google Scholar]
- Dami, I.; Hughes, H.G. Effects of PEG-induced water stress onin vitro hardening of ‘Valiant’grape. Plant Cell Tissue Organ Cult. 1997, 47, 97–101. [Google Scholar] [CrossRef]
- Lawlor, D. Absorption of polyethylene glycols by plants and their effects on plant growth. New Phytol. 1970, 69, 501–513. [Google Scholar] [CrossRef]
- Samarah, N.; Alqudah, A. Effects of late-terminal drought stress on seed germination and vigor of barley (Hordeum vulgare L.). Arch. Agron. Soil Sci. 2011, 57, 27–32. [Google Scholar] [CrossRef]
- Tabatabaei, S. Effect of osmo-priming on germination and enzyme activity in barley (Hordeum vulgare L.) seeds under drought stress conditions. J. Stress Physiol. Biochem. 2013, 9, 25–31. [Google Scholar]
- Abdel-Ghani, A.H.; Neumann, K.; Wabila, C.; Sharma, R.; Dhanagond, S.; Owais, S.J.; Börner, A.; Graner, A.; Kilian, B. Diversity of germination and seedling traits in a spring barley (Hordeum vulgare L.) collection under drought simulated conditions. Genet. Resour. Crop Evol. 2015, 62, 275–292. [Google Scholar] [CrossRef]
- Kaczmarek, M.; Fedorowicz-Strońska, O.; Głowacka, K.; Waśkiewicz, A.; Sadowski, J. CaCl2 treatment improves drought stress tolerance in barley (Hordeum vulgare L.). Acta Physiol. Plant. 2017, 39, 41. [Google Scholar] [CrossRef] [Green Version]
- Schmidthoffer, I.; Szilák, L.; Molnár, P.; Csontos, P.; Skribanek, A. Drought tolerance of European barley (Hordeum vulgare L.) varieties. Agriculture (Pol’nohospodárstvo) 2018, 64, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Chloupek, O.; Hrstkova, P.; Jurecka, D.; Graner, A. Tolerance of barley seed germination to cold- and drought-stress expressed as seed vigour. Plant Breed. 2003, 122, 199–203. [Google Scholar] [CrossRef]
- Hellal, F.; El-Shabrawi, H.; El-Hady, M.A.; Khatab, I.; El-Sayed, S.; Abdelly, C. Influence of PEG induced drought stress on molecular and biochemical constituents and seedling growth of Egyptian barley cultivars. J. Genet. Eng. Biotechnol. 2018, 16, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Amro, A.; Elakhdar, A.; Dawood, M.F.; Moursi, Y.S.; Baenziger, P.S. Marker-trait association for grain weight of spring barley in well-watered and drought environments. Mol. Biol. Rep. 2019, 46, 2907–2918. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falconer, D.S. Introduction to Quantitative Genetics; Pearson Education India: Bangalore, India, 1996. [Google Scholar]
- Sallam, A.; Amro, A.; Ammar, E.-A.; Dawood, M.F.; Kumamaru, T.; Baenziger, P.S. Genetic diversity and genetic variation in morpho-physiological traits to improve heat tolerance in Spring barley. Mol. Biol. Rep. 2018, 45, 2441–2453. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [Green Version]
- Sallam, A.; Mourad, A.M.; Hussain, W.; Baenziger, P.S. Genetic variation in drought tolerance at seedling stage and grain yield in low rainfall environments in wheat (Triticum aestivum L.). Euphytica 2018, 214, 169. [Google Scholar] [CrossRef]
- Oraguzie, N.C.; Gardiner, S.E.; Rikkerink, E.H.; Silva, H.N. Association Mapping in Plants; Springer: New York, NY, USA, 2007. [Google Scholar]
- Sallam, A.; Martsch, R. Association mapping for frost tolerance using multi-parent advanced generation inter-cross (MAGIC) population in faba bean (Vicia faba L.). Genetica 2015, 143, 501–514. [Google Scholar] [CrossRef]
- Naz, A.A.; Arifuzzaman, M.; Muzammil, S.; Pillen, K.; Léon, J. Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet. 2014, 15, 107. [Google Scholar] [CrossRef] [Green Version]
- Schrooten, C.; Bovenhuis, H. Detection of pleiotropic effects of quantitative trait loci in outbred populations using regression analysis. J. Dairy Sci. 2002, 85, 3503–3513. [Google Scholar] [CrossRef] [Green Version]
- Almodares, A.; Hadi, M.; Kharazian, Z.A. Sweet sorghum: Drought tolerance and high biomass sugar crop. In Biomass Detection Production and Usage; InTechOpen: Rijeka, Croatia, 2011. [Google Scholar]
- Hite, D.R.; Auh, C.; Scandalios, J.G. Catalase activity and hydrogen peroxide levels are inversely correlated in maize scutella during seed germination. Redox Rep. 1999, 4, 29–34. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvim, F.T.C.; Carolino, S.N.M.; Cascardo, J.C.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 2001, 126, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landi, S.; Capasso, G.; Azaiez, B.; Ezzahra, F.; Jallouli, S.; Ayadi, S.; Trifa, Y.; Esposito, S. Different roles of heat shock proteins (70 kDa) during abiotic stresses in barley (Hordeum vulgare) genotypes. Plants 2019, 8, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.-C.; Hsieh, E.-J.; Chen, J.-H.; Chen, H.-Y.; Lin, T.-P. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012, 158, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Wu, Z.; Cao, G.; Li, J.; Wei, J.; Tsuge, T.; Gu, H.; Aoyama, T.; Qu, L.-J. TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol. Plant 2014, 7, 601–615. [Google Scholar] [CrossRef] [Green Version]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV 1 transcription factor, phosphorylated by S n RK 2 kinases, regulates the expression of ABI 3, ABI 4, and ABI 5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Zhang, B.; Su, L.; Hu, B.; Li, L. Expression of AhDREB1, an AP2/ERF transcription factor gene from peanut, is affected by histone acetylation and increases abscisic acid sensitivity and tolerance to osmotic stress in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1441. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.-H.; Kim, D.-Y.; Shin, H.J.; Nam, K.J.; An, J.H.; Pack, I.-S.; Park, J.-H.; Jeong, S.-C.; Kim, H.B.; Kim, C.-G. Drought stress-induced compositional changes in tolerant transgenic rice and its wild type. Food Chem. 2014, 153, 145–150. [Google Scholar] [CrossRef]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.; Li, M.; Dong, Z.; Ji, H.; Li, X. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 2015, 34, 395–410. [Google Scholar] [CrossRef]
- Gachomo, E.W.; Jimenez-Lopez, J.C.; Baptiste, L.J.; Kotchoni, S.O. GIGANTUS1 (GTS1), a member of Transducin/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; He, F.; Tang, W.; Du, H.; Wang, H. Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes 2019, 10, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Priya, P.; Jain, M. Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta 2013, 238, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Ruan, M.-B.; Yang, Y.-L.; Li, K.-M.; Guo, X.; Wang, B.; Yu, X.-L.; Peng, M. Identification and characterization of drought-responsive CC-type glutaredoxins from cassava cultivars reveals their involvement in ABA signalling. BMC Plant Biol. 2018, 18, 329. [Google Scholar] [CrossRef]
- Kumar, A.; Dubey, A.K.; Kumar, V.; Ansari, M.A.; Narayan, S.; Kumar, S.; Pandey, V.; Shirke, P.A.; Pande, V.; Sanyal, I. Overexpression of rice glutaredoxin genes LOC_Os02g40500 and LOC_Os01g27140 regulate plant responses to drought stress. Ecotoxicol. Environ. Saf. 2020, 200, 110721. [Google Scholar] [CrossRef]
- Pan, C.; Tian, K.; Ban, Q.; Wang, L.; Sun, Q.; He, Y.; Yang, Y.; Pan, Y.; Li, Y.; Jiang, J. Genome-wide analysis of the biosynthesis and deactivation of gibberellin-dioxygenases gene family in Camellia sinensis (L.) O. Kuntze. Genes 2017, 8, 235. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.K.; Ryu, M.Y.; Seo, D.H.; Kang, B.G.; Kim, W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 2011, 157, 2240–2257. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Li, Y. Functional genomics of the protein kinase superfamily from wheat. Mol. Breed. 2019, 39, 141. [Google Scholar] [CrossRef]
- Waseem, M.; Li, Z. Dissecting the Role of a Basic Helix-Loop-Helix Transcription Factor, SlbHLH22, Under Salt and Drought Stresses in Transgenic Solanum lycopersicum L. Front. Plant Sci. 2019, 10, 734. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B. Roles of the Brassica napus DELLA Protein BnaA6. RGA, in Modulating Drought Tolerance by Interacting With the ABA Signaling Component BnaA10. ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef] [PubMed]
- Jarzyniak, K.M.; Jasiski, M. Membrane transporters and drought resistance-a complex issue. Front. Plant Sci. 2014, 5, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.Y.; Jin, J.Y.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 improves drought and drought stress resistance in Arabidopsis. Physiol. Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodger, J.Q.; Schachtman, D.P. Re-examining the role of ABA as the primary long-distance signal produced by water-stressed roots. Plant Signal. Behav. 2010, 5, 1298–1301. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Peng, Y.; Meckes, N.; Allen, S.; Stewart, C.N.; Traw, M.B. ATP-dependent binding cassette transporter G family member 16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 2014, 166, 879–888. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, R.; Smolka, U.; Altmann, S.; Eschen-Lippold, L.; Senning, M.; Sonnewald, S.; Weigel, B.; Frolova, N.; Strehmel, N.; Hause, G. The ABC transporter ABCG1 is required for suberin formation in potato tuber periderm. Plant Cell 2014, 26, 3403–3415. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Li, Y.; Ling, H.; Liu, S.; Liu, M.; Chen, J.; Guo, S. Transcriptomic analyses reveal clathrin-mediated endocytosis involved in symbiotic seed germination of Gastrodia elata. Bot. Stud. 2017, 58, 31. [Google Scholar] [CrossRef] [Green Version]
- Pagnussat, L.; Burbach, C.; Baluška, F.; de la Canal, L. Rapid endocytosis is triggered upon imbibition in Arabidopsis seeds. Plant Signal. Behav. 2012, 7, 416–421. [Google Scholar] [CrossRef] [Green Version]
- Haider, M.S.; Zhang, C.; Kurjogi, M.M.; Pervaiz, T.; Zheng, T.; Zhang, C.; Lide, C.; Shangguan, L.; Fang, J. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-Seq analysis. Sci. Rep. 2017, 7, 13134. [Google Scholar] [CrossRef] [Green Version]
- Callos, J.D.; DiRado, M.; Xu, B.; Behringer, F.J.; Link, B.M.; Medford, J.I. The forever young gene encodes an oxidoreductase required for proper development of the Arabidopsis vegetative shoot apex. Plant J. 1994, 6, 835–847. [Google Scholar] [CrossRef]
- Passaia, G.; Queval, G.; Bai, J.; Margis-Pinheiro, M.; Foyer, C.H. The effects of redox controls mediated by glutathione peroxidases on root architecture in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1403–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, L.; Li, X.; Chen, J.; Li, Y.; Tang, Y.; Lv, J. Photosynthetic and ascorbate-glutathione metabolism in the flag leaves as compared to spikes under drought stress of winter wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0194625. [Google Scholar] [CrossRef] [Green Version]
- Liting, W.; Lina, W.; Yang, Y.; Pengfei, W.; Tiancai, G.; Guozhang, K. Abscisic acid enhances tolerance of wheat seedlings to drought and regulates transcript levels of genes encoding ascorbate-glutathione biosynthesis. Front. Plant Sci. 2015, 6, 458. [Google Scholar]
- Elakhdar, A.; EL-Sattar, M.A.; Amer, K.; Rady, A.; Kumamaru, T. Population structure and marker–trait association of drought tolerance in barley (Hordeum vulgare L.). Comptes Rendus Biol. 2016, 339, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Hetz, W.; Hochholdinger, F.; Schwall, M.; Feix, G.n. Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J. 1996, 10, 845–857. [Google Scholar] [CrossRef]
- Utz, H. PLABSTAT: Ein Computerprogramm zur Statistischen Analyse von Pflanzenzüchterischen Experimenten, Version 2B; Institut für Pflanzenzüchtung, Saatgutforschung und Populationsgenetik, Universität Hohenheim: Stuttgart, Germany, 1997. [Google Scholar]
- Julkowska, M.M.; Saade, S.; Agarwal, G.; Gao, G.; Pailles, Y.; Morton, M.; Awlia, M.; Tester, M. MVAPP—Multivariate analysis application for streamlined data analysis and curation. Plant Physiol. 2019, 180, 1261–1276. [Google Scholar] [CrossRef] [Green Version]
- Poland, J.A.; Rife, T.W. Genotyping by sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Francis, D.; Merk, H.; Namuth-Covert, D. Introduction to Single Marker Analysis (SMA). 3–5. 2011. Available online: https://plant-breeding-genomics.extension.org/introduction-to-single-marker-analysis-sma/ (accessed on 26 August 2020).
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.R.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W. Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Abbreviation | Description of Measurement |
|---|---|---|
| Germination Percentage | G% | ×, where n is the number of germinated seeds at the end of the experiment, N is the total number of seeds. |
| Germination Pace | GP | where N is the number of germinated seeds at the end of the experiment, n is the number of newly germinated seed at certain day g, g = (1, 2, 3, …) |
| Root Length | RL | Root length was measured with a scaled ruler (in cm) |
| Shoot length | SL | Shoot length was measured with a scaled ruler (in cm) |
| Shoot–Root Length Ration | SRR | as the ratio of the SL to the RL |
| Fresh Weight | FW | Fresh weight was recorded in grams using a sensitive balance (Sartorius AC 1215, Germany) |
| Reduction of Germination Percentage | Reduction_G% | Reduction of G% = G% under control − G% under drought |
| Reduction of Germination Pace | Reduction_GP | Reduction of GP = GP under control − GP under drought |
| Reduction of Root Length | Reduction_RL | Reduction of RL = RL under control − RL under drought |
| Reduction of Shoot Length | Reduction_SL | Reduction of SL = SL under control − SL under drought |
| Reduction of Fresh Weight | Reduction_FW | Reduction of FW = FW under control − FW under drought |
| Drought Tolerance Index (Germination Percentage) | G%_DTI | |
| Drought Tolerance Index (Germination Pace) | GP_DTI | |
| Drought Tolerance Index (Root Length) | RL_DTI | |
| Drought Tolerance Index (Shoot Length) | SL_DTI | |
| Drought Tolerance Index (Fresh Weight) | FW_ DTI |
| Source of Variance | Germination Percentage | Germination Pace | Shoot Length | Root Length | Shoot/Root Ration | Fresh Weight |
|---|---|---|---|---|---|---|
| Treatments (T) | 118.77 ** | 115.76 ** | 591.64 ** | 467.03 ** | 333.09 ** | 29.58 ** |
| Replications | 1.76 | 5.64 ** | 0.36 | 0.01 | 3.29 * | 0.03 |
| Genotypes (G) | 14.55 ** | 9.19 ** | 23.92 ** | 43.05 ** | 57.57 ** | 26.83 ** |
| T × G | 10.83 ** | 5.19 ** | 15.57 ** | 20.46 ** | 12.30 ** | 12.60 ** |
| Genotype | Traits | G% | GP | SL | RL | FW |
|---|---|---|---|---|---|---|
| SCSAL-21 | 3 | × | × | × | ||
| PNBYT15 | 3 | × | × | × | ||
| SCSAL-36 | 3 | × | × | × | ||
| PNBYT1 | 3 | × | × | × | ||
| SC4-41 | 2 | × | × | |||
| SCBNB57 | 2 | × | × | |||
| SCSAL-52 | 2 | × | × | |||
| SCYT-28 | 2 | × | × | |||
| PNBYT27 | 2 | × | × | |||
| SC2-19 | 2 | × | × | |||
| INTROD-46 | 2 | × | × | |||
| SCSAL-10 | 2 | × | × | |||
| Giza135 | 2 | × | × |
| Treatment | No. of QTL | Chromosome. | R2 | |
|---|---|---|---|---|
| Traits | ||||
| Root Length Drought | C | - | - | - |
| D | 21 | 1, 2,3,4, 7 | 26.9–44.3% | |
| Shoot Length/Root Length Ratio Control | C | 1 | 7 | 34.3% |
| D | 2 | 1 | 52.7% | |
| Germination Percentage | C | 7 | 1, 2, 5 | 29.1–47.0% |
| D | 3 | 1, 2, 6 | 32.1–42.9% | |
| Germination Pace | C | 2 | 2, 5 | 38.8–46.1% |
| D | 5 | 3 | 35.7–43.9% | |
| Fresh Weight | C | 4 | 3, 6 | 27.5–55.1% |
| D | 4 | 3, 6 | 28.7–57.5% | |
| QTL under Control | 15 | |||
| QTL under Drought | 35 | |||
| Total | 50 | |||
| Drought tolerance index | ||||
| Fresh Weight Drought Tolerance Index | 1 | 1 | 34.2% | |
| Shoot Length Drought Tolerance Index | 1 | 7 | 36.2% | |
| Drought Tolerance Index | 3 | 6 | 30.9–33.1% | |
| Shoot Length Drought Tolerance Index | 11 | 1, 2, 7 | 28.0–58.2% | |
| Total | 16 | |||
| Reduction index | ||||
| Reduction of Fresh Weight | 1 | 3 | 47.6% | |
| Reduction of Shoot Length | 1 | 7 | 28.3% | |
| Reduction of Root Length | 1 | 5 | 37.8% | |
| Shoot Length/Root Length Ratio Drought | 1 | |||
| Fresh Weight Control | 1 | 2 | 30.2% | |
| Total | 5 | |||
| Total Number of QTLs for all traits | 71 | |||
| Marker | Chr | SNP Position | Traits | Candidate Genes | Annotation |
|---|---|---|---|---|---|
| S3_465596823 | 3H | 465596823 | FW_C FW_D | HORVU3Hr1G061080 HORVU3Hr1G061120 | Glutaredoxin family protein Gibberellin 2-beta-dioxygenase 1 |
| S3_471113003 | 3H | 471113003 | FW_C FW_D | HORVU3Hr1G061850 HORVU3Hr1G061860 | RING/U-box superfamily protein Protein kinase superfamily protein |
| S3_50419767 | 3H | 50419767 | FW_C FW_D | HORVU3Hr1G018980 HORVU3Hr1G019070 | Basic helix-loop-helix (bHLH) DNA-binding family protein Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family |
| S5_99278943 | 5H | 99278943 | RL_DTI Reduction_RL | HORVU5Hr1G021110 HORVU5Hr1G021120 | ABC transporter G family member 5 ABC transporter G family member 1 |
| S5_526418931 | 5H | 526418931 | G%_C GP_C | HORVU5Hr1G069950 | At4g40080-like protein |
| S6_15937118 | 6H | 15937118 | Reduction_G% G%_DTI | HORVU6Hr1G008640 HORVU6Hr1G008520 | Catalase 1 Chromosome 3B genomic scaffold |
| S6_15937146 | 6H | 15937146 | Reduction_G% G%_DTI | HORVU6Hr1G008730 HORVU6Hr1G008880 | Catalase 3 Heat shock 70 kDa protein C |
| S6_37139810 | 6H | 37139810 | FW_C FW_D | HORVU6Hr1G016350 HORVU6Hr1G016260 | Cytochrome P450 superfamily protein Transducin/WD40 repeat-like superfamily protein |
| S6_520541285 | 6H | 520541285 | Reduction_G% G%_DTI G%_D | HORVU6Hr1G075640 | AP2-like ethylene-responsive transcription factor |
| S7_61794629 | 7H | 61794629 | SL_DTI Reduction_SL | HORVU7Hr1G030700 HORVU7Hr1G030810 | Dehydrogenase/reductase SDR family member 4 Glutathione peroxidase 1 |
| QTL Name | Marker Allele | p-Value | R2 | Allele Eff 1 | Traits in This Study | Traits in Earlier Studies |
|---|---|---|---|---|---|---|
| DSI-GYPS-Gh | stm773-2_149 | 0.00010 | 22.9 | −0.433 | Red_FW | Drought susceptibility index for grain yield per spike (Sallam et al. 2019) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moursi, Y.S.; Thabet, S.G.; Amro, A.; Dawood, M.F.A.; Baenziger, P.S.; Sallam, A. Detailed Genetic Analysis for Identifying QTLs Associated with Drought Tolerance at Seed Germination and Seedling Stages in Barley. Plants 2020, 9, 1425. https://doi.org/10.3390/plants9111425
Moursi YS, Thabet SG, Amro A, Dawood MFA, Baenziger PS, Sallam A. Detailed Genetic Analysis for Identifying QTLs Associated with Drought Tolerance at Seed Germination and Seedling Stages in Barley. Plants. 2020; 9(11):1425. https://doi.org/10.3390/plants9111425
Chicago/Turabian StyleMoursi, Yasser S., Samar G. Thabet, Ahmed Amro, Mona F. A. Dawood, P. Stephen Baenziger, and Ahmed Sallam. 2020. "Detailed Genetic Analysis for Identifying QTLs Associated with Drought Tolerance at Seed Germination and Seedling Stages in Barley" Plants 9, no. 11: 1425. https://doi.org/10.3390/plants9111425
APA StyleMoursi, Y. S., Thabet, S. G., Amro, A., Dawood, M. F. A., Baenziger, P. S., & Sallam, A. (2020). Detailed Genetic Analysis for Identifying QTLs Associated with Drought Tolerance at Seed Germination and Seedling Stages in Barley. Plants, 9(11), 1425. https://doi.org/10.3390/plants9111425








