Thioredoxins: Emerging Players in the Regulation of Protein S-Nitrosation in Plants
Abstract
1. Introduction
2. Origin and Fate of Thiol S-Nitrosation in Plant Cells
3. S-Nitrosoglutathione Reductase Indirectly Regulates Protein S-Nitrosation Status in Plants
4. The Key Role of the Mammalian Thioredoxin System in Protein Denitrosation
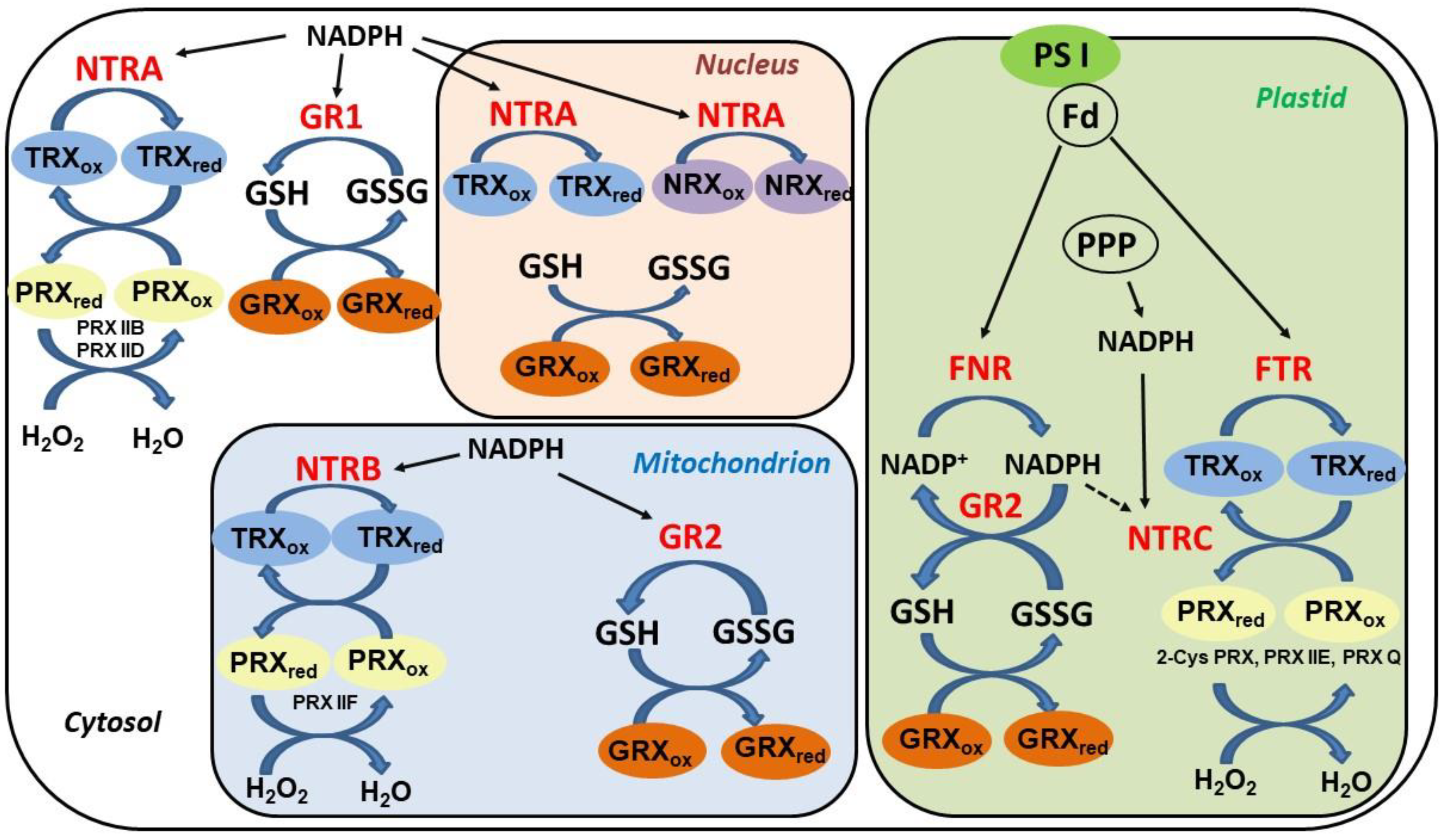
5. Thioredoxin Systems in Higher Plants
6. TRX Role in Denitrosation of Plant Proteins
7. Complementary Role of Plant Peroxiredoxins and Sulfiredoxins in the TRX-Dependent Denitrosation
8. Other Mechanisms of Protein Denitrosation in Plants
9. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Walther, D. The Roles of Post-translational Modifications in the Context of Protein Interaction Networks. PLoS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef] [PubMed]
- Friso, G.; Van Wijk, K.J. Update: Post-translational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, T.; Chen, W.; Oka, S.-I.; Fu, C.; Jain, M.R.; Parrott, A.M.; Baykal, A.T.; Sadoshima, J.; Li, H. Redox Regulatory Mechanism of Transnitrosylation by Thioredoxin. Mol. Cell. Proteom. 2010, 9, 2262–2275. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Komatsu, S. Posttranslational Modifications and Plant–Environment Interaction. Methods Enzymol. 2017, 586, 97–113. [Google Scholar] [PubMed]
- Spoel, S.H. Orchestrating the proteome with post-translational modifications. J. Exp. Bot. 2018, 69, 4499–4503. [Google Scholar] [CrossRef] [PubMed]
- Klomsiri, C.; Karplus, P.A.; Poole, L.B. Cysteine-Based Redox Switches in Enzymes. Antioxid. Redox Signal. 2011, 14, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef]
- Waszczak, C.; Akter, S.; Jacques, S.; Huang, J.; Messens, J.; Van Breusegem, F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 2015, 66, 2923–2934. [Google Scholar] [CrossRef]
- Ruiz-May, E.; Segura-Cabrera, A.; Elizalde-Contreras, J.M.; Shannon, L.M.; Loyola-Vargas, V.M. A recent advance in the intracellular and extracellular redox post-translational modification of proteins in plants. J. Mol. Recognit. 2018, 32, e2754. [Google Scholar] [CrossRef]
- Šírová, J.; Sedlářová, M.; Piterková, J.; Luhová, L.; Petrivalsky, M. The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 2011, 181, 560–572. [Google Scholar] [CrossRef]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2017, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Del Río, L.A.; Palma, J.M. Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes. Plants 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Neill, S.J. Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds. Plants 2019, 8, 41. [Google Scholar] [CrossRef]
- Petřivalský, M.; Luhová, L. Nitrated Nucleotides: New Players in Signaling Pathways of Reactive Nitrogen and Oxygen Species in Plants. Front. Plant Sci. 2020, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Lindermayr, C. Nitric Oxide-Dependent Posttranslational Modification in Plants: An Update. Int. J. Mol. Sci. 2012, 13, 15193–15208. [Google Scholar] [CrossRef]
- Kovacs, I.; Lindermayr, C. Nitric oxide-based protein modification: Formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 2013, 4, 137. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; Padilla, M.N.; Barroso, J.B. S-nitrosothiols function during abiotic stress in plants. J. Exp. Bot. 2019, 70, 4429–4439. [Google Scholar] [CrossRef]
- Umbreen, S.; Lubega, J.; Loake, G.J. Sulphur: The heart of nitric oxide-dependent redox signalling. J. Exp. Bot. 2019, 70, 4279–4286. [Google Scholar] [CrossRef]
- Kolbert, Z.; Barroso, J.; Brouquisse, R.; Corpas, F.; Gupta, K.; Lindermayr, C.; Loake, G.; Palma, J.; Petřivalský, M.; Wendehenne, D.; et al. A forty year journey: The generation and roles of NO in plants. Nitric Oxide 2019, 93, 53–70. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T.; et al. Regulating the regulator: Nitric oxide control of post-translational modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Stamler, J.S. Regulation byS-Nitrosylation of Protein Post-translational Modification. J. Biol. Chem. 2011, 287, 4411–4418. [Google Scholar] [CrossRef] [PubMed]
- Seth, D.; Stamler, J.S. The SNO-proteome: Causation and classifications. Curr. Opin. Chem. Biol. 2011, 15, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Sánchez-Calvo, B.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 2018, 69, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Palma, J.M.; Del Rio, L.A.; Barroso, J.B. Evidence supporting the existence of l-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009, 184, 9–14. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M. Assessing Nitric Oxide (NO) in Higher Plants: An Outline. Nitrogen 2018, 1, 12–20. [Google Scholar] [CrossRef]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; LaMattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.-S.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase–like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2. [Google Scholar] [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef]
- Hill, B.G.; Dranka, B.P.; Bailey, S.M.; Lancaster, J.R., Jr.; Darley-Usmar, V.M. What Part of NO Don’t You Understand? Some Answers to the Cardinal Questions in Nitric Oxide Biology. J. Biol. Chem. 2010, 285, 19699–19704. [Google Scholar] [CrossRef]
- Heinrich, T.A.; Da Silva, R.S.; Miranda, K.M.; Switzer, C.H.; Wink, D.A.; Fukuto, J.M. Biological nitric oxide signalling: Chemistry and terminology. Br. J. Pharmacol. 2013, 169, 1417–1429. [Google Scholar] [CrossRef]
- Elamotte, O.; Bertoldo, J.B.; Besson-Bard, A.; Erosnoblet, C.; Aimé, S.; Ehichami, S.; Terenzi, H.; Wendehenne, D. Protein S-nitrosylation: Specificity and identification strategies in plants. Front. Chem. 2015, 2, 114. [Google Scholar]
- Gaston, B. Nitric oxide and thiol groups. Biochim. Biophys. Acta 1999, 1411, 323–333. [Google Scholar] [CrossRef]
- Handy, D.E.; Loscalzo, J. Nitric Oxide and Posttranslational Modification of the Vascular Proteome. Arter. Thromb. Vasc. Biol. 2006, 26, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Kuruthukulangarakoola, G.T.; Lindermayr, C. Regulation and Function of Protein S-Nitrosylation in Plant Stress. In Stress Signaling in Plants: Genomics and Proteomics Perspective; Sarwat, M., Ahmad, A., Abdin, M., Eds.; Springer: New York, NY, USA, 2013; Volume 1. [Google Scholar]
- Mata-Pérez, C.; Spoel, S.H. Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 2019, 279, 27–33. [Google Scholar] [CrossRef]
- Yun, B.-W.; Skelly, M.J.; Yin, M.; Yu, M.; Mun, B.; Lee, S.; Hussain, A.; Spoel, S.H.; Loake, G.J. Nitric oxide and S -nitrosoglutathione function additively during plant immunity. New Phytol. 2016, 211, 516–526. [Google Scholar] [CrossRef]
- Martínez-Ruiz, A. S-nitrosylation: A potential new paradigm in signal transduction. Cardiovasc. Res. 2004, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Alché, J.D.D.; Barroso-Albarracín, J.B. Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 2013, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.I.; Hussain, A.; Yun, B.-W.; Spoel, S.H.; Loake, G.J. GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci. 2011, 181, 540–544. [Google Scholar] [CrossRef]
- Broniowska, K.A.; Hogg, N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal. 2012, 17, 969–980. [Google Scholar] [CrossRef]
- Lancaster, J.R., Jr. Protein cysteine thiol nitrosation: Maker or marker of reactive nitrogen species-induced nonerythroid cellular signaling? Nitric Oxide 2008, 19, 68–72. [Google Scholar] [CrossRef]
- Li, Q.; Lancaster, J.R. A Conspectus of Cellular Mechanisms of Nitrosothiol Formation from Nitric Oxide. Forum Immunopathol. Dis. Ther. 2012, 3, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.R. How are nitrosothiols formed de novo in vivo ? Arch. Biochem. Biophys. 2017, 617, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Lipton, S.A. Emerging Role of Protein-Protein Transnitrosylation in Cell Signaling Pathways. Antioxid. Redox Signal. 2013, 18, 239–249. [Google Scholar] [CrossRef]
- Seth, D.; Hess, D.T.; Hausladen, A.; Wang, L.; Wang, Y.-J.; Stamler, J.S. A Multiplex Enzymatic Machinery for Cellular Protein S-nitrosylation. Mol. Cell 2018, 69, 451–464.e6. [Google Scholar] [CrossRef] [PubMed]
- Wolhuter, K.; Whitwell, H.J.; Switzer, C.H.; Burgoyne, J.R.; Timms, J.F.; Eaton, P. Evidence against Stable Protein S-Nitrosylation as a Widespread Mechanism of Post-translational Regulation. Mol. Cell 2018, 69, 438–450.e5. [Google Scholar] [CrossRef]
- Clementi, E.; Brown, G.C.; Feelisch, M.; Moncada, S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. USA 1998, 95, 7631–7636. [Google Scholar] [CrossRef]
- Benhar, M.; Forrester, M.T.; Stamler, J.S. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 721–732. [Google Scholar] [CrossRef]
- Paige, J.S.; Xu, G.; Stancevic, B.; Jaffrey, S.R. Nitrosothiol Reactivity Profiling Identifies S-Nitrosylated Proteins with Unexpected Stability. Chem. Biol. 2008, 15, 1307–1316. [Google Scholar] [CrossRef]
- Liu, L.; Hausladen, A.; Zeng, M.; Que, L.; Heitman, J.; Stamler, J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nat. Cell Biol. 2001, 410, 490–494. [Google Scholar] [CrossRef]
- Barnett, S.D.; Buxton, I.L.O. The role of S-nitrosoglutathione reductase (GSNOR) in human disease and therapy. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 340–354. [Google Scholar] [CrossRef]
- Jahnová, J.; Luhová, L.; Petrivalsky, M. S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 2019, 8, 48. [Google Scholar] [CrossRef]
- Kubienová, L.; Kopečný, D.; Tylichová, M.; Briozzo, P.; Skopalová, J.; Šebela, M.; Navrátil, M.; Tâche, R.; Luhová, L.; Barroso, J.B.; et al. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie 2013, 95, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Guerra, D.; Lee, U.; Vierling, E. S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 2013, 4, 430. [Google Scholar] [CrossRef] [PubMed]
- Guerra, D.; Ballard, K.; Truebridge, I.; Vierling, E. S-Nitrosation of Conserved Cysteines Modulates Activity and Stability ofS-Nitrosoglutathione Reductase (GSNOR). Biochemie 2016, 55, 2452–2464. [Google Scholar] [CrossRef]
- Kovacs, I.; Holzmeister, C.; Wirtz, M.; Geerlof, A.; Fröhlich, T.; Römling, G.; Kuruthukulangarakoola, G.T.; Linster, E.; Hell, R.; Arnold, G.J.; et al. ROS-Mediated Inhibition of S-nitrosoglutathione Reductase Contributes to the Activation of Anti-oxidative Mechanisms. Front. Plant Sci. 2016, 7, 1669. [Google Scholar] [CrossRef] [PubMed]
- Tichá, T.; Lochman, J.; Činčalová, L.; Luhová, L.; Petřivalský, M. Redox regulation of plant S-nitrosoglutathione reductase activity through post-translational modifications of cysteine residues. Biochem. Biophys. Res. Commun. 2017, 494, 27–33. [Google Scholar] [CrossRef]
- Tichá, T.; Činčalová, L.; Kopečný, D.; Sedlářová, M.; Kopečná, M.; Luhová, L.; Petřivalský, M. Characterization of S-nitrosoglutathione reductase from Brassica and Lactuca spp. and its modulation during plant development. Nitric Oxide 2017, 68, 68–76. [Google Scholar] [CrossRef]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef]
- Kubienová, L.; Tichá, T.; Jahnová, J.; Luhová, L.; Mieslerová, B.; Petřivalský, M. Effect of abiotic stress stimuli on S-nitrosoglutathione reductase in plants. Planta 2013, 239, 139–146. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free. Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Rouhier, N.; Cerveau, D.; Couturier, J.; Reichheld, J.-P.; Rey, P. Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 2015, 1850, 1479–1496. [Google Scholar] [CrossRef]
- Hondal, R.J.; Ruggles, E.L. Differing views of the role of selenium in thioredoxin reductase. Amino Acids 2011, 41, 73–89. [Google Scholar] [CrossRef]
- Nikitovic, D.; Holmgren, A. S-Nitrosoglutathione Is Cleaved by the Thioredoxin System with Liberation of Glutathione and Redox Regulating Nitric Oxide. J. Biol. Chem. 1996, 271, 19180–19185. [Google Scholar] [CrossRef] [PubMed]
- Benhar, M.; Forrester, M.T.; Hess, U.T.; Stamler, J.S. Regulated Protein Denitrosylation by Cytosolic and Mitochondrial Thioredoxins. Science 2008, 320, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Ben-Lulu, S.; Ziv, T.; Admon, A.; Weisman-Shomer, P.; Benhar, M. A Substrate Trapping Approach Identifies Proteins Regulated by Reversible S-nitrosylation. Mol. Cell. Proteom. 2014, 13, 2573–2583. [Google Scholar] [CrossRef] [PubMed]
- Stoyanovsky, D.A.; Tyurina, Y.Y.; Tyurin, V.A.; Anand, D.; Mandavia, D.N.; Gius, D.; Ivanova, J.; Pitt, B.; Billiar, T.R.; Kagan, V.E. Thioredoxin and Lipoic Acid Catalyze the Denitrosation of Low Molecular Weight and ProteinS-Nitrosothiols. J. Am. Chem. Soc. 2005, 127, 15815–15823. [Google Scholar] [CrossRef]
- Sengupta, R.; Ryter, S.W.; Zuckerbraun, B.S.; Tzeng, E.; Billiar, T.R.; Stoyanovsky, D.A. Thioredoxin Catalyzes the Denitrosation of Low-Molecular Mass and ProteinS-Nitrosothiols. Biochemistry 2007, 46, 8472–8483. [Google Scholar] [CrossRef]
- Barglow, K.T.; Knutson, C.G.F.; Wishnok, J.S.; Tannenbaum, S.R.; Marletta, M.A. Site-specific and redox-controlled S-nitrosation of thioredoxin. Proc. Natl. Acad. Sci. USA 2011, 108, E600–E606. [Google Scholar] [CrossRef]
- Sengupta, R.; Holmgren, A. Thioredoxin and Thioredoxin Reductase in Relation to Reversible S-Nitrosylation. Antioxid. Redox Signal. 2013, 18, 259–269. [Google Scholar] [CrossRef]
- Engelman, R.; Ziv, T.; Arnér, E.S.J.; Benhar, M. Inhibitory nitrosylation of mammalian thioredoxin reductase 1: Molecular characterization and evidence for its functional role in cellular nitroso-redox imbalance. Free Radic. Biol. Med. 2016, 97, 375–385. [Google Scholar] [CrossRef]
- Benhar, M. Roles of mammalian glutathione peroxidase and thioredoxin reductase enzymes in the cellular response to nitrosative stress. Free Radic. Biol. Med. 2018, 127, 160–164. [Google Scholar] [CrossRef]
- Espinosa, B.; Arnér, E.S.J. Thioredoxin-related protein of 14 kDa as a modulator of redox signalling pathways. Br. J. Pharmacol. 2018, 176, 544–553. [Google Scholar] [CrossRef]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.-P.; Riondet, C. Thioredoxin and Glutaredoxin Systems in Plants: Molecular Mechanisms, Crosstalks, and Functional Significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Dos Santos, C.V.; Rey, P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006, 11, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.-P.; Khafif, M.; Riondet, C.; Droux, M.; Bonnard, G.; Meyer, Y. Inactivation of Thioredoxin Reductases Reveals a Complex Interplay between Thioredoxin and Glutathione Pathways in Arabidopsis Development. Plant Cell 2007, 19, 1851–1865. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Barman, D.N.; Kim, M.G.; Kim, W.-Y. Stress defense mechanisms of NADPH-dependent thioredoxin reductases (NTRs) in plants. Plant Signal. Behav. 2015, 10, e1017698. [Google Scholar] [CrossRef]
- Delorme-Hinoux, V.; Bangash, S.A.; Meyer, A.J.; Reichheld, J.-P. Nuclear thiol redox systems in plants. Plant Sci. 2016, 243, 84–95. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Foresi, N.; Delledonne, M.; LaMattina, L. Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J. Exp. Bot. 2013, 64, 3339–3349. [Google Scholar] [CrossRef]
- Berger, H.; De Mia, M.; Morisse, S.; Marchand, C.H.; Lemaire, S.D.; Wobbe, L.; Kruse, O. A Light Switch Based on Protein S-Nitrosylation Fine-Tunes Photosynthetic Light Harvesting in Chlamydomonas1. Plant Physiol. 2016, 171, 821–832. [Google Scholar]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Kneeshaw, S.; Gelineau, S.; Tada, Y.; Loake, G.J.; Spoel, S.H. Selective Protein Denitrosylation Activity of Thioredoxin-h5 Modulates Plant Immunity. Mol. Cell 2014, 56, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.-P. Interplay between the NADP-Linked Thioredoxin and Glutathione Systems in Arabidopsis Auxin Signaling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Cejudo, F.J.; LaMattina, L. Nitric oxide is required for the auxin-induced activation of NADPH-dependent thioredoxin reductase and protein denitrosylation during root growth responses in arabidopsis. Ann. Bot. 2015, 116, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Ghanta, S.; Bhattacharyya, D.; Sinha, R.; Banerjee, A.; Chattopadhyay, S. Nicotiana tabacum overexpressing γ-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta 2011, 233, 895–910. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Morisse, S.; Bedhomme, M.; Marchand, C.H.; Festa, M.; Rouhier, N.; Lemaire, S.D.; Trost, P. Mechanisms of Nitrosylation and Denitrosylation of Cytoplasmic Glyceraldehyde-3-phosphate Dehydrogenase fromArabidopsis thaliana. J. Biol. Chem. 2013, 288, 22777–22789. [Google Scholar] [CrossRef]
- Zhao, Y.; He, M.; Ding, J.; Xi, Q.; Loake, G.J.; Zheng, W. Regulation of Anticancer Styrylpyrone Biosynthesis in the Medicinal Mushroom Inonotus obliquus Requires Thioredoxin Mediated Transnitrosylation of S-nitrosoglutathione Reductase. Sci. Rep. 2016, 6, 37601. [Google Scholar] [CrossRef]
- Liebthal, M.; Maynard, D.; Dietz, K.-J. Peroxiredoxins and Redox Signaling in Plants. Antioxid. Redox Signal. 2018, 28, 609–624. [Google Scholar] [CrossRef]
- Dietz, K.-J. Peroxiredoxins in Plants and Cyanobacteria. Antioxid. Redox Signal. 2011, 15, 1129–1159. [Google Scholar] [CrossRef]
- Sakamoto, A.; Tsukamoto, S.; Yamamoto, H.; Ueda-Hashimoto, M.; Takahashi, M.; Suzuki, H.; Morikawa, H. Functional complementation in yeast reveals a protective role of chloroplast 2-Cys peroxiredoxin against reactive nitrogen species. Plant J. 2003, 33, 841–851. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic Identification of S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Laxa, M.; Mattè, A.; Zaninotto, F.; Finkemeier, I.; Jones, A.M.; Perazzolli, M.; Vandelle, E.; Dietz, K.-J.; Delledonne, M. S-Nitrosylation of Peroxiredoxin II E Promotes Peroxynitrite-Mediated Tyrosine Nitration. Plant Cell 2007, 19, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Engelman, R.; Weisman-Shomer, P.; Ziv, T.; Xu, J.; Arnér, E.S.J.; Benhar, M. Multilevel Regulation of 2-Cys Peroxiredoxin Reaction Cycle byS-Nitrosylation. J. Biol. Chem. 2013, 288, 11312–11324. [Google Scholar] [CrossRef]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-dependent reduction of cysteine–sulphinic acid by S. cerevisiae sulphiredoxin. Nat. Cell Biol. 2003, 425, 980–984. [Google Scholar] [CrossRef]
- Sunico, C.R.; Sultan, A.; Nakamura, T.; Dolatabadi, N.; Parker, J.; Shan, B.; Han, X.; Yates, J.R.; Masliah, E.; Ambasudhan, R.; et al. Role of sulfiredoxin as a peroxiredoxin-2 denitrosylase in human iPSC-derived dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2016, 113, E7564–E7571. [Google Scholar] [CrossRef]
- Rey, P.; Bécuwe, N.; Barrault, M.-B.; Rumeau, D.; Havaux, M.; Biteau, B.; Toledano, M.B. The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. Plant J. 2007, 49, 505–514. [Google Scholar] [CrossRef]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef]
- Root, P.; Sliskovic, I.; Mutus, B. Platelet cell-surface protein disulphide-isomerase mediated S-nitrosoglutathione consumption. Biochem. J. 2004, 382, 575–580. [Google Scholar] [CrossRef][Green Version]
- Sliskovic, I.; Raturi, A.; Mutus, B. Characterization of the S-Denitrosation Activity of Protein Disulfide Isomerase. J. Biol. Chem. 2004, 280, 8733–8741. [Google Scholar] [CrossRef]
- Houston, N.L.; Fan, C.; Xiang, J.Q.; Schulze, J.-M.; Jung, R.; Boston, R.S. Phylogenetic Analyses Identify 10 Classes of the Protein Disulfide Isomerase Family in Plants, Including Single-Domain Protein Disulfide Isomerase-Related Proteins. Plant Physiol. 2005, 137, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, X.; Li, R.; Yuan, L.; Dai, Y.; Wang, X. Identification and Functional Analysis of a Protein Disulfide Isomerase (AtPDI1) in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 913. [Google Scholar] [CrossRef]
- Jourd’Heuila, D.; Larouxa, F.S.; Miles, A.M.; Wink, D.A.; Grisham, M.B. Effect of Superoxide Dismutase on the Stability ofS-Nitrosothiols. Arch. Biochem. Biophys. 1999, 361, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Macdonald, T.L.; Mannick, J.B.; Conaway, M.R.; Gaston, B. AcceleratedS-Nitrosothiol Breakdown by Amyotrophic Lateral Sclerosis Mutant Copper,Zinc-Superoxide Dismutase. J. Biol. Chem. 2001, 276, 39872–39878. [Google Scholar] [CrossRef] [PubMed]
- Schonhoff, C.M.; Matsuoka, M.; Tummala, H.; Johnson, M.A.; Estevéz, A.G.; Wu, R.; Kamaid, A.; Ricart, K.C.; Hashimoto, Y.; Gaston, B.; et al. S-nitrosothiol depletion in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2006, 103, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [PubMed]
- Emengel, A.; Echaki, M.; Eshekariesfahlan, A.; Lindermayr, C. Effect of nitric oxide on gene transcription—S-nitrosylation of nuclear proteins. Front. Plant Sci. 2013, 4, 293. [Google Scholar]
- Spyrou, G.; Enmark, E.; Miranda-Vizuete, A.; Gustafsson, J. Cloning and Expression of a Novel Mammalian Thioredoxin. J. Biol. Chem. 1997, 272, 2936–2941. [Google Scholar] [CrossRef]
- Kneeshaw, S.; Spoel, S.H. Thioredoxin-Dependent Decomposition of Protein S-Nitrosothiols. In Nitric Oxide. Methods in Molecular Biology; Mengel, A., Lindermayr, C., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1747. [Google Scholar]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jedelská, T.; Luhová, L.; Petřivalský, M. Thioredoxins: Emerging Players in the Regulation of Protein S-Nitrosation in Plants. Plants 2020, 9, 1426. https://doi.org/10.3390/plants9111426
Jedelská T, Luhová L, Petřivalský M. Thioredoxins: Emerging Players in the Regulation of Protein S-Nitrosation in Plants. Plants. 2020; 9(11):1426. https://doi.org/10.3390/plants9111426
Chicago/Turabian StyleJedelská, Tereza, Lenka Luhová, and Marek Petřivalský. 2020. "Thioredoxins: Emerging Players in the Regulation of Protein S-Nitrosation in Plants" Plants 9, no. 11: 1426. https://doi.org/10.3390/plants9111426
APA StyleJedelská, T., Luhová, L., & Petřivalský, M. (2020). Thioredoxins: Emerging Players in the Regulation of Protein S-Nitrosation in Plants. Plants, 9(11), 1426. https://doi.org/10.3390/plants9111426





