Short De-Etiolation Increases the Rooting of VC801 Avocado Rootstock
Abstract
1. Introduction
2. Results
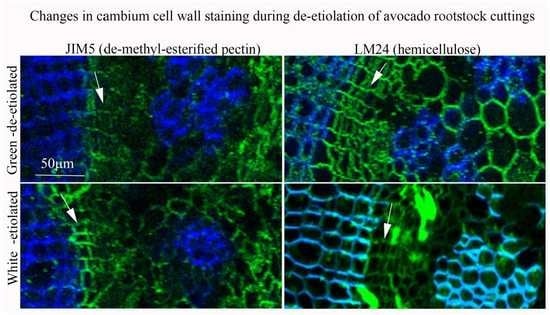
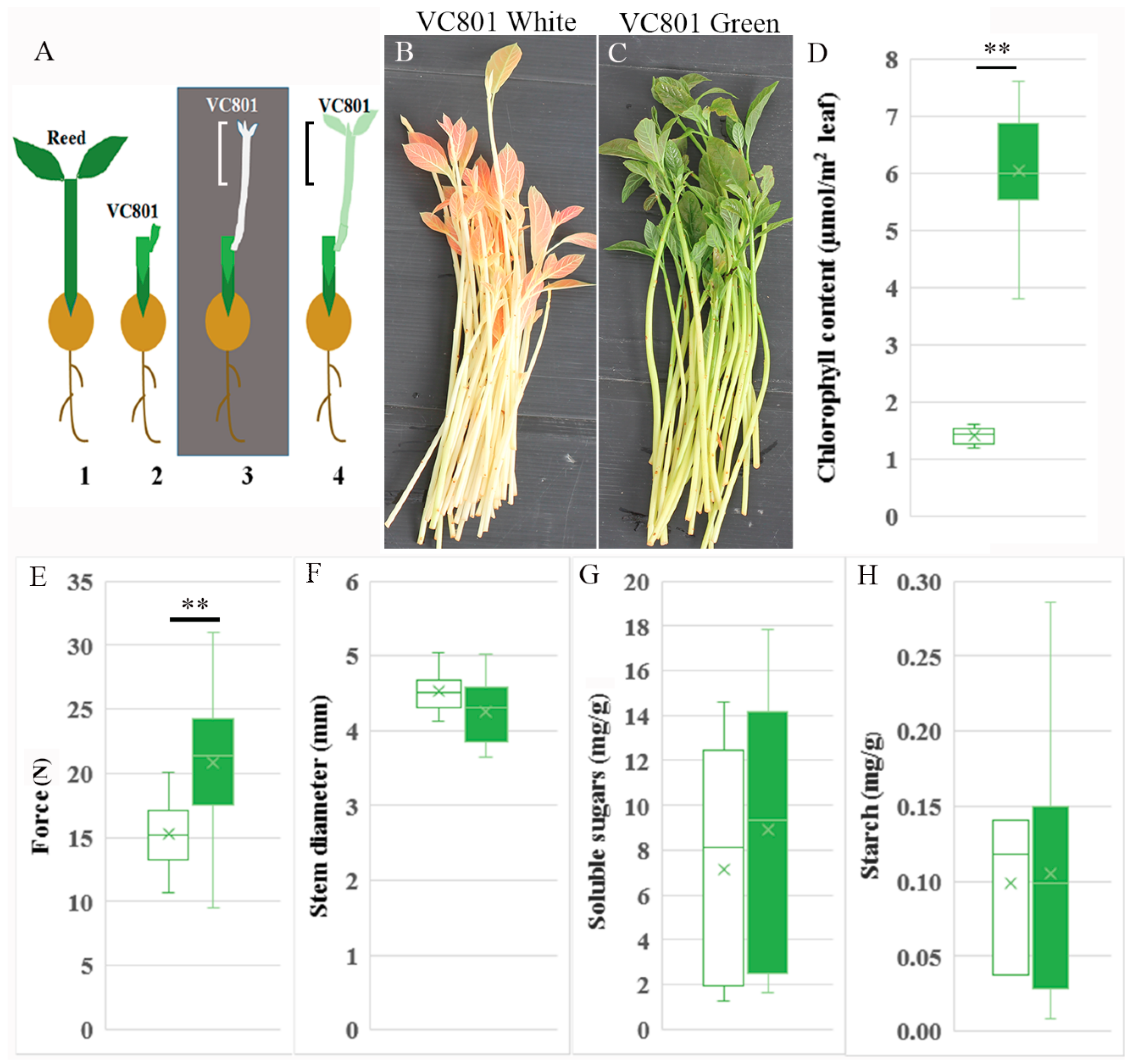
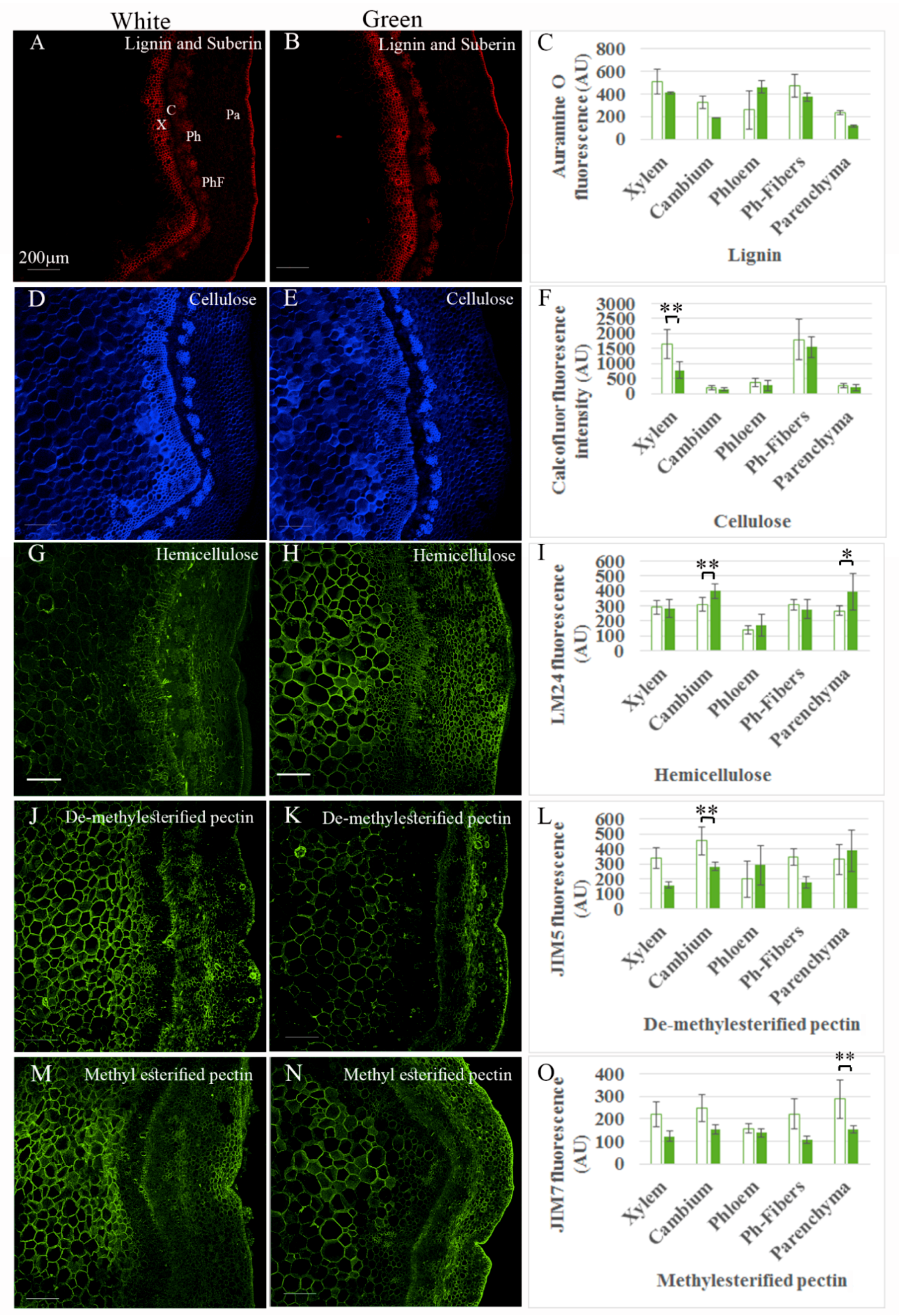
2.1. Etiolated VC801 Avocado Cuttings Are Softer and Differ in Their Tissue and Cell-Wall Composition from De-Etiolated Branches
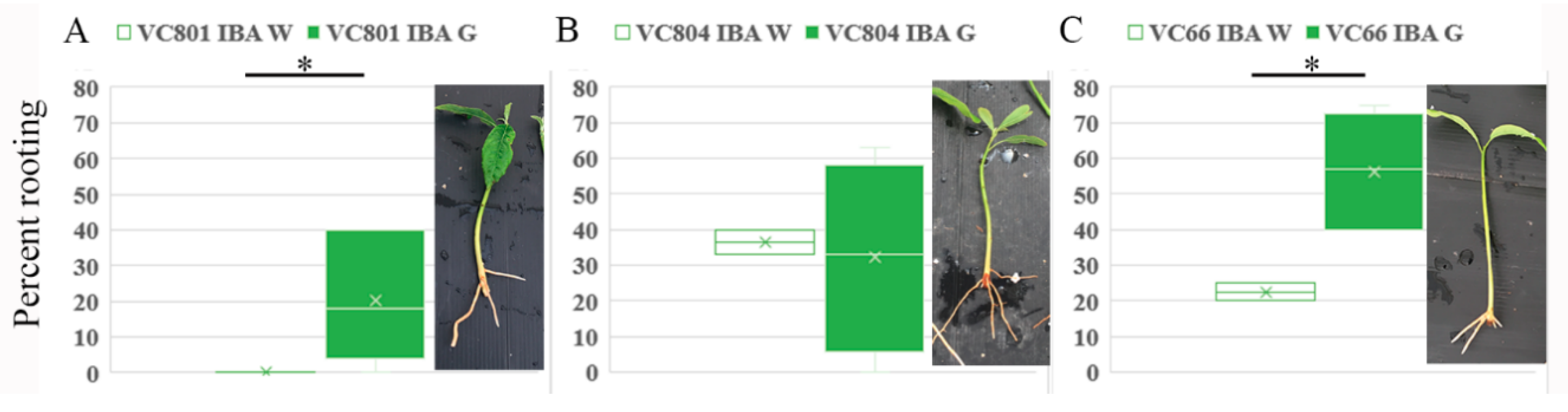
2.2. Induction of Adventitious Root Formation in Etiolated and De-Etiolated Branches of VC801
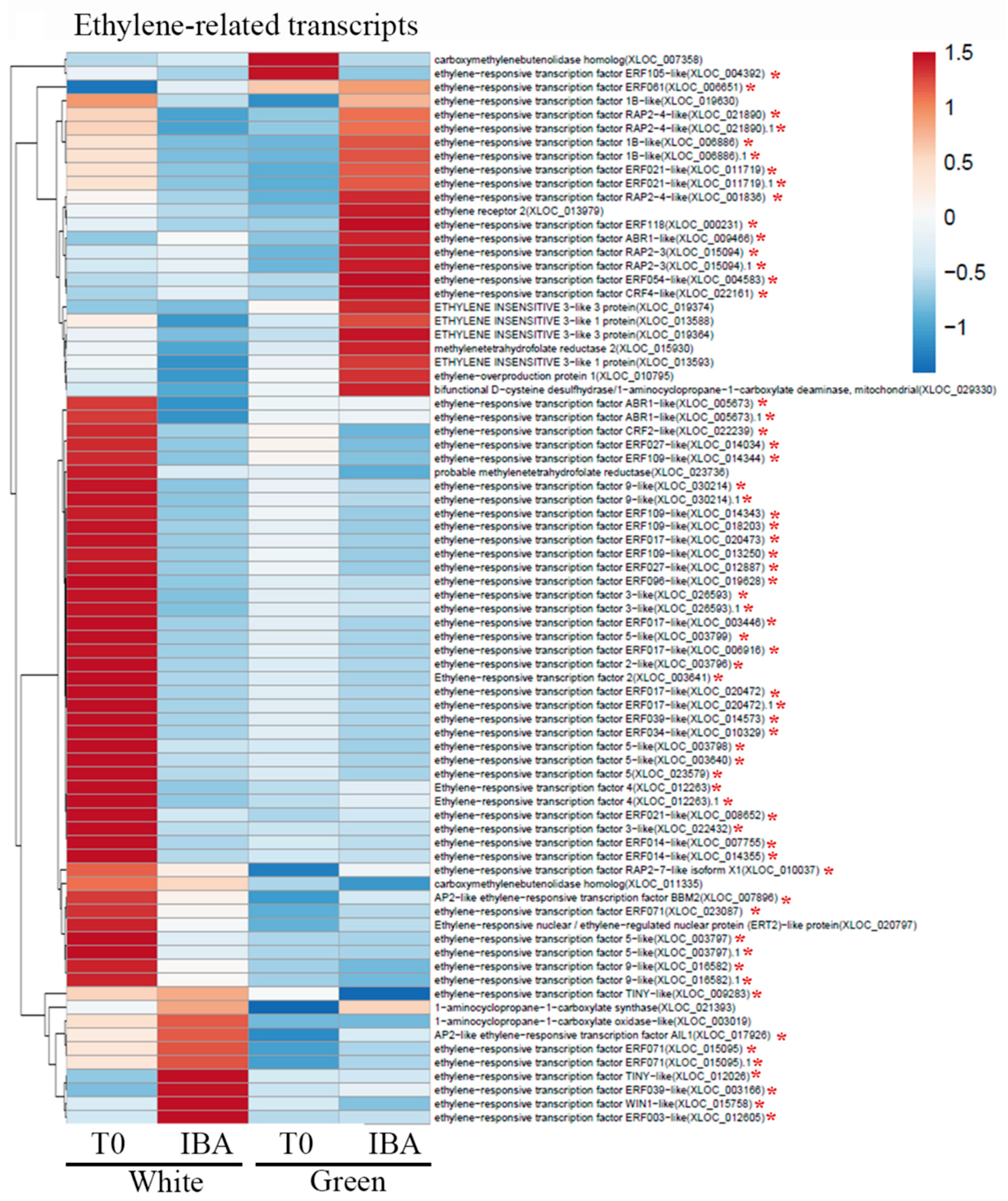
2.3. Hormone and Transcript Profiles of VC801 Etiolated and De-Etiolated Branches
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Fluorescent Staining
4.3. Confocal Microscopy and Image Analysis
4.4. Induction of Adventitious Roots
4.5. RNA Isolation, RNA Sequencing and Bioinformatics
4.6. Compression Analysis
4.7. Chlorophyll Measurements
4.8. Auxin Measurements
4.9. Sugar Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Galindo-Tovar, M.E.; Ogata-Aguilar, N.; Arzate-Fernandez, A.M. Some aspects of avocado (Persea americana Mill.) diversity and domestication in Mesoamerica. Genet. Resour. Crop Evol. 2008, 55, 441–450. [Google Scholar] [CrossRef]
- Melnyk, C.W.; Meyerowitz, E.M. Plant grafting. Curr. Biol. 2015, 25, R183–R188. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ya’acov, A.; Michelson, E. Avocado Rootstocks. Horticultural Reviews; Janick, J., Ed.; John Wiley and Sons, Inc.: New York, NY, USA, 1995; Volume 17, pp. 381–429. [Google Scholar]
- Smith, L.A.; Dann, E.K.; Pegg, K.G.; Whiley, A.W.; Giblin, F.R.; Doogan, V.; Kopittke, R. Field assessment of avocado rootstock selections for resistance to Phytophthora root rot. Aust. Plant Pathol. 2011, 40, 39–47. [Google Scholar] [CrossRef]
- De Villiers, A.; Ernst, A. Avocado rootstock research: Principels and practices. World Avocado Congr. 2015, 8, 40–45. [Google Scholar]
- Ben-Ya′acov, A.; Zilberstaine, M. The possible use of avocado (Persea americana mill.) germplasm material as rootstocks for soil stress conditions. Rev. Chapingo Ser. Hortic. 1999, 5, 25–28. [Google Scholar]
- Ben-Ya’acov, A.; Michelson, E.; Zilberstaine, M.; Barkan, Z.; Sela, I. Selection of Clonal Avocado Rootstocks in Israel for High Productivity under Different Soil Conditions. In Proceedings of the 2nd World Avocado Congress, Orange, CA, USA, 21–26 April 1992; pp. 521–526. [Google Scholar]
- Ben-Ya’acov, A.; Zilberstaine, M. Clonal avocado (Persea americana Mill.) rootstocks in Israel. Rev. Chapingo Ser. Hortic. 1999, 5, 39–42. [Google Scholar]
- Lazare, S.; Haberman, A.; Yermiyahu, U.; Erel, R.; Simenski, E.; Dag, A. Avocado rootstock influences scion leaf mineral content. Arch. Agron. Soil Sci. 2019, 1–11. [Google Scholar] [CrossRef]
- Haberman, A.; Tsror Lahkim, L.; Lazare, S.; Hazanovsky, M.; Lebiush, S.; Zipori, I.; Busatn, A.; Simenski, E.; Dag, A. Management of Verticillium Wilt of Avocado Using Tolerant Rootstocks. Plants 2020, 9, 531. [Google Scholar] [CrossRef] [PubMed]
- Frolich, E.F.; Platt, R.G. Use of the etiolation technique in rooting avocado cuttings. Calif. Avocado Soc. Yearbook 1972, 55, 97–109. [Google Scholar]
- Brokaw, W.H. Rootrot resistant avocado clonal rootstocks. Plant Prop. 1975, 21, 7–8. [Google Scholar]
- Ernst, A.A.; Holtzhausen, L.C. New promising technique for rooting difficult-to-root avocado (Persea americana Mill.) cuttings. Citrus Sub-Trop. Fruit J. 1978, 532, 6–10. [Google Scholar]
- Goren, M. Approaches to the Production of Avocado Cuttings with Etiolated Bases, and the Endogenous Changes Involved in the Rooting Process. Master’s Thesis, Faculty of Agriculture of the Hebrew University of Jerusalem, Jerusalem, Israel, 1982. [Google Scholar]
- Hartmann, H.T.; Kester, D.E.; Davies, F.T.J.; Geneve, R.L. Plant Propagation. Principles and Practices, 9th ed.; Pearson Education Limited: Essex, UK, 2011. [Google Scholar]
- Ahsan, M.U.; Hayward, A.; Alam, M.; Bandaralage, J.H.; Topp, B.; Beveridge, C.A.; Mitter, N. Scion control of miRNA abundance and tree maturity in grafted avocado. BMC Plant Biol. 2019, 19, 382. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. MiRNA Control of Vegetative Phase Change in Trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 727. [Google Scholar] [CrossRef]
- Wulf, K.E.; Reid, J.B.; Foo, E. Auxin transport and stem vascular reconnection—Has our thinking become canalized? Ann. Bot. 2019, 123, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Damari-Weissler, H.; Rachamilevitch, S.; Aloni, R.; German, M.A.; Cohen, S.; Zwieniecki, M.A.; Michele Holbrook, N.; Granot, D. LeFRK2 is required for phloem and xylem differentiation and the transport of both sugar and water. Planta 2009, 230, 795–805. [Google Scholar] [CrossRef]
- Foster, T.M.; McAtee, P.A.; Waite, C.N.; Boldingh, H.L.; McGhie, T.K. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hort. Res. 2017, 4, 17009. [Google Scholar] [CrossRef]
- Moing, A.; Salesses, G.; Saglio, P.H. Growth and the composition and transport of carbohydrate in compatible and incompatible peach/plum grafts. Tree Physiol. 1987, 3, 345–354. [Google Scholar] [CrossRef]
- Agulló-Antón, M.Á.; Sánchez-Bravo, J.; Acosta, M.; Druege, U. Auxins or Sugars: What Makes the Difference in the Adventitious Rooting of Stored Carnation Cuttings? J. Plant Growth Reg. 2011, 30, 100–113. [Google Scholar] [CrossRef]
- Koukourikou-Petridou, M.A. Etiolation of stock plants affects adventitious root formation and hormone content of pea stem cuttings. Plant Growth Reg. 1998, 25, 17–21. [Google Scholar] [CrossRef]
- Sorin, C.; Bussell, J.D.; Camus, I.; Ljung, K.; Kowalczyk, M.; Geiss, G.; McKhann, H.; Garcion, C.; Vaucheret, H.; Sandberg, G.; et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 2005, 17, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, M.; Krens, F.A.; Visser, R.G.F.; De Klerk, G.-J.M. Etiolation and flooding of donor plants enhance the capability of Arabidopsis explants to root. Plant Cell Tissue Organ Cult. 2017, 130, 531–541. [Google Scholar] [CrossRef]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Klopotek, Y.; Haensch, K.T.; Hause, B.; Hajirezaei, M.R.; Druege, U. Dark exposure of petunia cuttings strongly improves adventitious root formation and enhances carbohydrate availability during rooting in the light. J. Plant Physiol. 2010, 167, 547–554. [Google Scholar] [CrossRef]
- Seluzicki, A.; Burko, Y.; Chory, J. Dancing in the dark: Darkness as a signal in plants. Plant Cell Environ. 2017, 40, 2487–2501. [Google Scholar] [CrossRef]
- Maynard, B.K.; Bassuk, N.B. Effects of Stock Plant Etiolation, Shading, Banding, and Shoot Development on Histology and Cutting Propagation of Carpinus betulus L. fastigiata. J. Amer. Hort. Sci. 1996, 121, 853–860. [Google Scholar] [CrossRef]
- Rogers, L.A.; Campbell, M.M. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004, 164, 17–30. [Google Scholar] [CrossRef]
- Fahn, A. Plant Anatomy, 4th ed.; Pergamon Press: Oxford, UK, 1990. [Google Scholar]
- Lev-Yadun, S. Plant development: Cell movement relative to each other is both common and very important. Plant Signal. Behav. 2015, 10, e991566. [Google Scholar] [CrossRef]
- Bhalerao, R.P.; Fischer, U. Environmental and hormonal control of cambial stem cell dynamics. J. Exp. Bot. 2017, 68, 79–87. [Google Scholar] [CrossRef]
- Ludwig-Muller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef]
- Rendon-Anaya, M.; Ibarra-Laclette, E.; Mendez-Bravo, A.; Lan, T.; Zheng, C.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacon-Lopez, A.; Hernandez-Guzman, G.; Chang, T.H.; et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Shi, H.; Xue, C.; Wei, N.; Guo, H.; Deng, X.W. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 2014, 111, 3913–3920. [Google Scholar] [CrossRef]
- Lakehal, A.; Chaabouni, S.; Cavel, E.; Le Hir, R.; Ranjan, A.; Raneshan, Z.; Novak, O.; Pacurar, D.I.; Perrone, I.; Jobert, F.; et al. A Molecular Framework for the Control of Adventitious Rooting by the TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis. Mol. Plant 2019. [Google Scholar] [CrossRef]
- Verstraeten, I.; Schotte, S.; Geelen, D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 2014, 5, 495. [Google Scholar] [CrossRef]
- Forlani, S.; Masiero, S.; Mizzotti, C. Fruit ripening: The role of hormones, cell wall modifications and their intersection with pathogens. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Merelo, P.; Agusti, J.; Arbona, V.; Costa, M.L.; Estornell, L.H.; Gomez-Cadenas, A.; Coimbra, S.; Gomez, M.D.; Perez-Amador, M.A.; Domingo, C.; et al. Cell Wall Remodeling in Abscission Zone Cells during Ethylene-Promoted Fruit Abscission in Citrus. Front. Plant Sci. 2017, 8, 126. [Google Scholar] [CrossRef]
- Xiao, C.; Somerville, C.; Anderson, C.T. Polygalacturonase involved in expansion1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 2014, 26, 1018–1035. [Google Scholar] [CrossRef] [PubMed]
- Duman, Z.; Eliyahu, A.; Abu-Abied, M.; Sadot, E. The contribution of cell wall remodeling and signaling to lateral organs formation. Isr. J. Plant Sci. 2020, 67, 110. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Dommes, J.; Gaspar, T.; D’Angeli, S.; Falasca, G.; Altamura, M.M. Rooting blockage in the tobacco rac mutant occurs at the initiation phase, and induces diversion to xylem differentiation. Plant Biosys. 2003, 137, 163–174. [Google Scholar] [CrossRef]
- Strader, L.C.; Culler, A.H.; Cohen, J.D.; Bartel, B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010, 153, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Yoder, A.; Bartel, B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 2000, 156, 1323–1337. [Google Scholar]
- Abu-Abied, M.; Szwerdszarf, D.; Mordehaev, I.; Levy, A.; Stelmakh, O.R.; Belausov, E.; Yaniv, Y.; Uliel, S.; Katzenellenbogen, M.; Riov, J.; et al. Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of Eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. Plant J. 2012, 71, 787–799. [Google Scholar] [CrossRef]
- Rasmussen, A.; Hosseini, S.A.; Hajirezaei, M.R.; Druege, U.; Geelen, D. Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J. Exp. Bot. 2015, 66, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.; San-Jose, M.C.; Vidal, N.; Fernandez-Lorenzo, J.L.; Vieitez, A.M. Anatomical and biochemical events during in vitro rooting of microcuttings from juvenile and mature phases of chestnut. Ann. Bot. 1999, 83, 619–629. [Google Scholar] [CrossRef]
- Diaz-Sala, C.; Hutchison, K.W.; Goldfarb, B.; Greenwood, M.S. Maturation-related loss in rooting competence by loblolly pine stem cuttings: The role of auxin transport, metabolism and tissue sensitivity. Physiol. Plant. 1996, 97, 481–490. [Google Scholar] [CrossRef]
- Abarca, D.; Pizarro, A.; Hernandez, I.; Sanchez, C.; Solana, S.P.; Del Amo, A.; Carneros, E.; Diaz-Sala, C. The GRAS gene family in pine: Transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots. BMC Plant Biol. 2014, 14, 354. [Google Scholar] [CrossRef]
- Eliyahu, A.; Duman, Z.; Sherf, S.; Genin, O.; Cinnamon, Y.; Abu-Abied, M.; Weinstain, R.; Dag, A.; Sadot, E. Vegetative propagation of elite Eucalyptus clones as food source for honeybees (Apis mellifera); adventitious roots versus callus formation. Isr. J. Plant Sci. 2020, 67, 83. [Google Scholar] [CrossRef]
- Sachs, T. Cell polarity and tissue patterning in plants. Development 1991, 113, 83–93. [Google Scholar]
- Sachs, T.; Cohen, D. Circular Vessels and the Control of Vascular Differentiation in Plants. Differentiation 1982, 21, 22–26. [Google Scholar] [CrossRef]
- Sauer, M.; Balla, J.; Luschnig, C.; Wisniewska, J.; Reinohl, V.; Friml, J.; Benkova, E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006, 20, 2902–2911. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Macedo, E.; Vieira, C.; Carrizo, D.; Porfirio, S.; Hegewald, H.; Arnholdt-Schmitt, B.; Calado, M.L.; Peixe, A. Adventitious root formation in olive (Olea europaea L.) microshoots: Anatomical evaluation and associated biochemical changes in peroxidase and polyphenol oxidase activities. J. Hort. Sci. Biotechnol. 2013, 88, 53–59. [Google Scholar] [CrossRef]
- Porfirio, S.; Calado, M.L.; Noceda, C.; Cabrita, M.J.; da Silva, M.G.; Azadi, P.; Peixe, A. Tracking biochemical changes during adventitious root formation in olive (Olea europaea L.). Sci. Hort. 2016, 204, 41–53. [Google Scholar] [CrossRef]
- Santos Macedo, E.; Sircar, D.; Cardoso, H.G.; Peixe, A.; Arnholdt-Schmitt, B. Involvement of alternative oxidase (AOX) in adventitious rooting of Olea europaea L. microshoots is linked to adaptive phenylpropanoid and lignin metabolism. Plant Cell Rep. 2012, 31, 1581–1590. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- de María, N.; Guevara, M.Á.; Perdiguero, P.; Vélez, M.D.; Cabezas, J.A.; López-Hinojosa, M.; Li, Z.; Díaz, L.M.; Pizarro, A.; Mancha, J.A.; et al. Molecular study of drought response in the Mediterranean conifer Pinus pinaster Ait.: Differential transcriptomic profiling reveals constitutive water deficit-independent drought tolerance mechanisms. Ecol. Evol. 2020, 10, 9788–9807. [Google Scholar] [CrossRef] [PubMed]
- Assefa, T.A.; Vandesompele, J.; Thas, O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020, 21, 312. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Lorbiecke, R.; Sauter, M. Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol. 1999, 119, 21–30. [Google Scholar] [CrossRef]
- Negi, S.; Sukumar, P.; Liu, X.; Cohen, J.D.; Muday, G.K. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 2010, 61, 3–15. [Google Scholar] [CrossRef]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 2019, 165, 90–100. [Google Scholar] [CrossRef]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorska, R.; Beeckman, T.; Friml, J.; Benkova, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef]
- Swarup, R.; Perry, P.; Hagenbeek, D.; Van Der Straeten, D.; Beemster, G.T.; Sandberg, G.; Bhalerao, R.; Ljung, K.; Bennett, M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 2007, 19, 2186–2196. [Google Scholar] [CrossRef]
- Kumpf, R.P.; Shi, C.L.; Larrieu, A.; Sto, I.M.; Butenko, M.A.; Peret, B.; Riiser, E.S.; Bennett, M.J.; Aalen, R.B. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 2013, 110, 5235–5240. [Google Scholar] [CrossRef]
- Huang, W.N.; Liu, H.K.; Zhang, H.H.; Chen, Z.; Guo, Y.D.; Kang, Y.F. Ethylene-induced changes in lignification and cell wall-degrading enzymes in the roots of mungbean (Vigna radiata) sprouts. Plant Physiol. Biochem. 2013, 73, 412–419. [Google Scholar] [CrossRef]
- Armezzani, A.; Abad, U.; Ali, O.; Andres Robin, A.; Vachez, L.; Larrieu, A.; Mellerowicz, E.J.; Taconnat, L.; Battu, V.; Stanislas, T.; et al. Transcriptional induction of cell wall remodelling genes is coupled to microtubule-driven growth isotropy at the shoot apex in Arabidopsis. Development 2018, 145, dev162255. [Google Scholar] [CrossRef] [PubMed]
- Faik, A.; Price, N.J.; Raikhel, N.V.; Keegstra, K. An Arabidopsis gene encoding an α-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 7797–7802. [Google Scholar] [CrossRef]
- Minic, Z.; Rihouey, C.; Do, C.T.; Lerouge, P.; Jouanin, L. Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol. 2004, 135, 867–878. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, W.; Sechet, J.; Martin, M.; Bovio, S.; Lionnet, C.; Long, Y.; Battu, V.; Mouille, G.; Monéger, F.; et al. Xyloglucans and Microtubules Synergistically Maintain Meristem Geometry and Phyllotaxis. Plant Physiol. 2019, 181, 1191–1206. [Google Scholar] [CrossRef]
- Vilches-Barro, A.; Maizel, A. Talking through walls: Mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2015, 23, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Hofte, H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef]
- Guenin, S.; Mareck, A.; Rayon, C.; Lamour, R.; Assoumou Ndong, Y.; Domon, J.M.; Senechal, F.; Fournet, F.; Jamet, E.; Canut, H.; et al. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 2011, 192, 114–126. [Google Scholar] [CrossRef]
- Kurihara, D.; Mizuta, Y.; Sato, Y.; Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 2015, 142, 4168–4179. [Google Scholar] [CrossRef] [PubMed]
- Ursache, R.; Andersen, T.G.; Marhavy, P.; Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018, 93, 399–412. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Warren, G.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Lugassi, N.; Kelly, G.; Arad, T.; Farkash, C.; Yaniv, Y.; Yeselson, Y.; Schaffer, A.A.; Raveh, E.; Granot, D.; Carmi, N. Expression of Hexokinase in Stomata of Citrus Fruit Reduces Fruit Transpiration and Affects Seed Development. Front. Plant Sci. 2020, 11, 255. [Google Scholar] [CrossRef]
- Goren, S.; Lugassi, N.; Stein, O.; Yeselson, Y.; Schaffer, A.A.; David-Schwartz, R.; Granot, D. Suppression of sucrose synthase affects auxin signaling and leaf morphology in tomato. PLoS ONE 2017, 12, e0182334. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duman, Z.; Hadas-Brandwein, G.; Eliyahu, A.; Belausov, E.; Abu-Abied, M.; Yeselson, Y.; Faigenboim, A.; Lichter, A.; Irihimovitch, V.; Sadot, E. Short De-Etiolation Increases the Rooting of VC801 Avocado Rootstock. Plants 2020, 9, 1481. https://doi.org/10.3390/plants9111481
Duman Z, Hadas-Brandwein G, Eliyahu A, Belausov E, Abu-Abied M, Yeselson Y, Faigenboim A, Lichter A, Irihimovitch V, Sadot E. Short De-Etiolation Increases the Rooting of VC801 Avocado Rootstock. Plants. 2020; 9(11):1481. https://doi.org/10.3390/plants9111481
Chicago/Turabian StyleDuman, Zvi, Gal Hadas-Brandwein, Avi Eliyahu, Eduard Belausov, Mohamad Abu-Abied, Yelena Yeselson, Adi Faigenboim, Amnon Lichter, Vered Irihimovitch, and Einat Sadot. 2020. "Short De-Etiolation Increases the Rooting of VC801 Avocado Rootstock" Plants 9, no. 11: 1481. https://doi.org/10.3390/plants9111481
APA StyleDuman, Z., Hadas-Brandwein, G., Eliyahu, A., Belausov, E., Abu-Abied, M., Yeselson, Y., Faigenboim, A., Lichter, A., Irihimovitch, V., & Sadot, E. (2020). Short De-Etiolation Increases the Rooting of VC801 Avocado Rootstock. Plants, 9(11), 1481. https://doi.org/10.3390/plants9111481