Abstract
The relationship between air pollution and the allergenic capacity of pollen is widely accepted, with allergenicity being directly related to air pollution. To our knowledge, this is the first study comparing the differential expression of Lolium perenne pollen genes by RNAseq, in two wild populations with different levels of air pollution. The objective is to search for proteins that are expressed differentially in both situations and to establish a relationship with increased allergenic capacity. Two populations of L. perenne (Madrid and Ciudad Real) have been studied in two consecutive years, under the rationale that overexpressed genes in Madrid, with higher levels of NO2 and SO2, could be a cause for their greater allergenic capacity. Heat shock proteins (HSP), glycoside hydrolases, proteins with leucin-rich repeat motifs, and proteins with EF-HAND motifs were consistently overexpressed in Madrid pollen in the two years studied. Interestingly, some genes were overexpressed only in one of the years studied, such as pectinesterases in the first year, and lipid transfer proteins (LTPs) and thaumatin in the second. Despite the fact that the potential of all these proteins in relation to possible allergies has been reported, this is the first time they are cited as possible allergens of L. perenne. The results found can contribute decisively to the knowledge of the allergens of L. perenne and their relationship with atmospheric pollution, and to the development of much more effective vaccines.
1. Introduction
Most immunoglobulin E (IgE)-mediated allergies are caused by the plant’s allergens, which may cause different symptoms such as rhinoconjunctivitis, edema, urticarial, asthma, and anaphylaxis [1]. Nowadays, the incidence of pollen allergy is undergoing a striking increase, with pollen allergens being the main cause among people with perennial allergic rhinitis [2,3]. Allergies and hypersensitive responses that are initiated by specific immunologic mechanisms triggered by pollen and other allergens, constitute one of the major health issues in modern societies [4].
The relationship between air pollution and the increased allergenic capacity of pollen is a widely accepted fact [5,6,7,8,9,10,11]. The relationship between the allergenic capacity of pollen, the degree of air pollution, and the physiological status of the plant has been recently demonstrated [12], revealing that plants growing under a higher atmospheric pollution had a lower photosynthetic efficiency, with altered ROS scavenging systems resulting in a greater degree of oxidative stress, higher H2O2 concentration, and enhanced NADPH oxidase activity in pollen. These two factors (H2O2 concentration and NADPH oxidase) are considered as very relevant in the increase of the allergenic capacity of pollen [12].
Pollen from different types of plants has been found to trigger allergic reactions [13]. Up to eleven groups of grass pollen allergens have been identified for its ability to elicit a specific IgE response [14]. Among them, pollen of the Poaceae family are the main source allergens as outstanding Poa pratensis (Kentucky bluegrass), Phleum pratense, and L. perenne (perennial ryegrass). [15]. The pollen of perennial ryegrass is the major cause of allergic diseases [16,17,18].
On the other hand, according to clinical data collected by allergists consultations through the usual skin tests, the true extent of the problem is not evidenced, as there are many more cases of pollen allergies. To date, six pollen allergens of L. perenne have been described and it is assumed that they are always present in pollen of this species, regardless of the place of collection. This is certainly one of the problems, since pollen used in skin tests rarely comes from the place where the patient lives. Furthermore, it is necessary to investigate new allergens, since in many cases vaccines do not have the desired effect, probably because they are not being manufactured with the appropriate allergens (Dr. F. Feo and Dra. T. Alfaya personal communication).
Plants have an inducible adaptative metabolism known as secondary metabolism, through which they are able to adapt to both biotic and abiotic stresses. This metabolism is capable of synthesizing a plethora of molecules of different nature that only appear under stress [19]. The best known and most studied are molecules related to the defense against pathogens. Some of them have demonstrated their allergenic capacity in some plant’s pollen, such as lipid transfer proteins (PR 14) or thaumatin (PR 5) [20]. As discussed above, the relationship between air pollution and the greater allergenic capacity of some pollen has already been demonstrated, which is surely related to the synthesis of molecules induced by these environmental conditions, probably allergenic molecules not yet described.
Pollen used in this work was collected from two cities in central Spain (Madrid and Ciudad Real), near heavy traffic areas in both cases. However, pollution levels, especially nitrogen oxides and sulfur oxides, were higher in Madrid throughout the plant’s vegetative and reproductive period [12]. The pollen was collected from the plants before leaving the anthers.
Once the relationship of air pollution with a greater allergenic capacity of pollen had been demonstrated, the objective of this work was to identify overexpressed genes in the high pollution area (Madrid) as compared to the low pollution area (Ciudad Real, considered as control) by means of a transcriptomic analysis, with the secondary aim to find new potential allergens in the pollen of L. perenne. In order to achieve these objectives, the gene differential expression of pollen from both localities was studied using the RNAseq technique, in two consecutive years, placing a special emphasis on those genes that were overexpressed in the Madrid pollen, and that could be responsible for its greater allergenic capacity.
2. Results
To construct a transcriptome database, six mRNA libraries were generated in each collection moment (May 2017 and 2018) by Illumina sequencing, three from each population of L. perenne from Ciudad Real and three from Madrid.
Table 1 summarizes the mapping results. Among the 52 million readings (on average with 101 bp read length) obtained in each sample, 53% of them were mapped. Uniformity between the samples, indicated that samples were comparable. Usually, it is possible to align 60–90% of the reads to the reference genome. However, this data depends upon the quality of the sample and the coverage of the reference genome. The highest percentages are obtained with very well-curated model organism genomes.

Table 1.
Total, mapped, and HQ (High Quality) reads of the three different replicates from pollen from Madrid and Ciudad Real in the two sampling moments, 2017 and 2018.
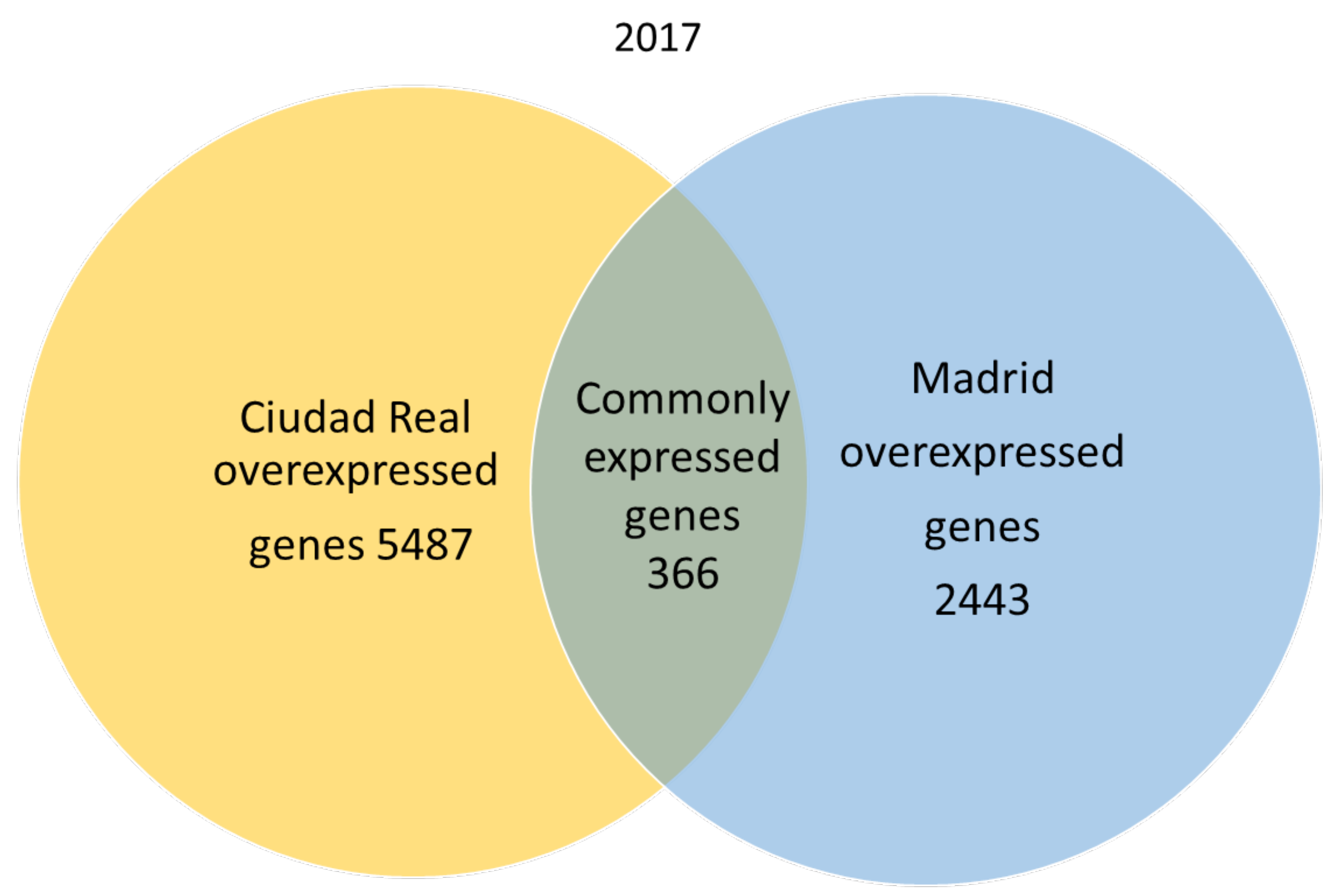
A total of 666,180 genes were identified in 2017 after sequencing, mapping alignment, normalized expression, and differential expression, but 550,285 genes in 2017 were not available for the study, since the fold change and p adjust value were not obtained in the normalized expression analysis. A total of 115,895 genes were available for the genetic expression analysis, of which 107,599 had a p adjust value over 0.05. When the pollen of the two cities was compared, the expression pattern showed that in 2017, 366 genes were common to both cities (expression without significant differences), and 2443 genes were significantly overexpressed in the pollen from Madrid, while 5487 genes were significantly overexpressed in Ciudad Real (Figure 1).
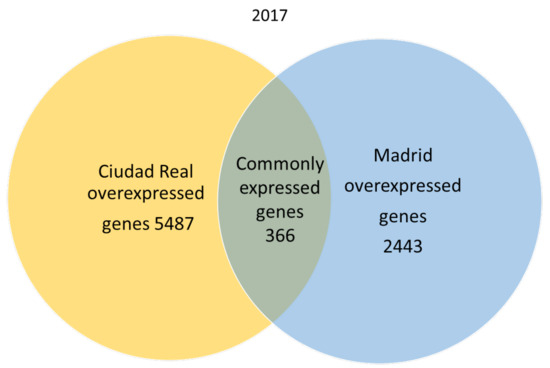
Figure 1.
Venn diagram of overexpressed and common genes in the samples of pollen from Madrid and Ciudad Real in 2017 with a p adjust value < 0.05.
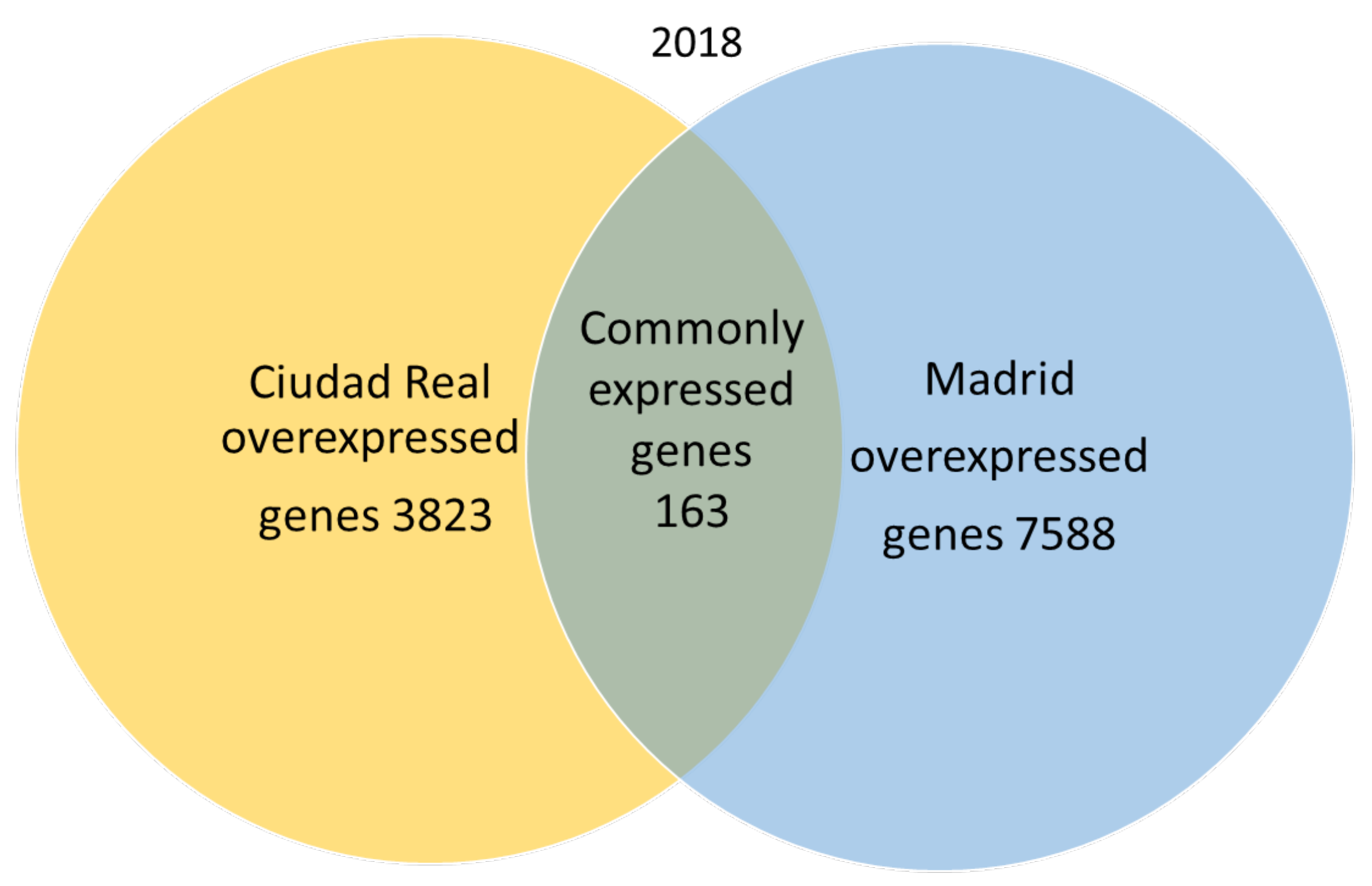
A total of 666,181 genes were identified in 2018 after sequencing, mapping alignment, normalized expression, and differential expression, but 585,368 genes were not available for the study, since the fold change and p adjust value were not obtained in the normalized expression analysis. A total of 80,813 genes resulted as available for the genetic expression analysis, of which 69,239 had a p adjust value over 0.05. When the pollen of the two cities was compared, the expression pattern showed that in 2018, 163 genes were common to both cities (expression without significant differences), and 7568 genes were significantly overexpressed in the pollen from Madrid, while 3823 genes were significantly overexpressed in Ciudad Real (Figure 2).
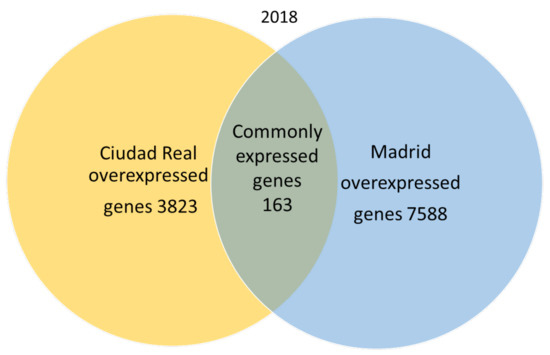
Figure 2.
Venn diagram of overexpressed and common genes in the samples of pollen from Madrid and Ciudad Real in 2018 with a p adjust value < 0.05.
Table 2 shows the genes that have been identified with some description in any of the Gene Ontology (GO), KEGG Orthology (KO), Pfam, and Clusters of Orthologous Groups (COG) databases, among the 50 most overexpressed (based in the fold change value) in Madrid and Ciudad Real in both years.

Table 2.
Among the 50 genes with the highest differential expression (based in the fold change value), those that have some description in the databases used are indicated in this table: GO (Gene Ontology), KO (KEGG Orthology), Pfam, and COG (Clusters of Orthologous Groups). (a) Genes overexpressed in Madrid in 2017; (b) genes overexpressed in Ciudad Real in 2017; (c) genes overexpressed in Madrid in 2018; (d) genes overexpressed in Ciudad Real in 2018. The rest of the genes can be found in the Supplementary Materials.
In both years, the highest number of genes identified corresponded to genes overexpressed in the pollen from Madrid, eight out of 50 in 2017 and 25 out of 50 in 2018 compared to two and eight in Ciudad Real, respectively.
In Ciudad Real, overexpressed genes were related to the primary metabolism, none of which was involved in routes of secondary adaptive metabolism by which potential allergenic molecules were synthesized. Conversely, among the genes overexpressed in the Madrid pollen some were involved in secondary metabolism, especially in 2018. In 2017, genes involved in the shikimic acid pathway, in the phenylpropanoid pathway, and heat shock proteins of family 20 (HSP 20) were overexpressed. In 2018, almost all overexpressed genes were related to secondary metabolism. There were 14 isoforms of the heat shock protein of 20 family, two isoforms of allergen 1 from Betula verrucosa (Bet v 1), and two lipid transport proteins (LTPs). All of them have been described as allergens.
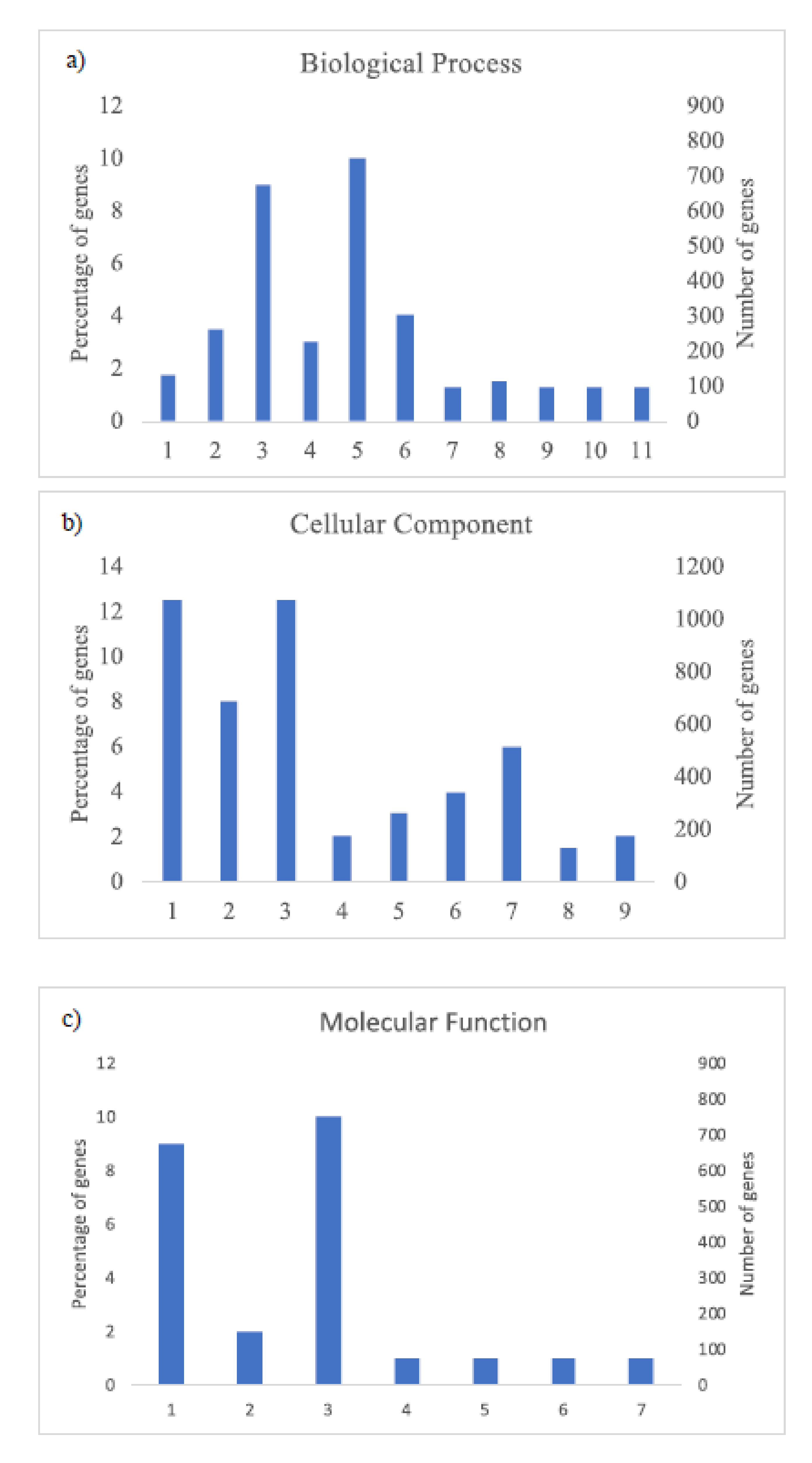
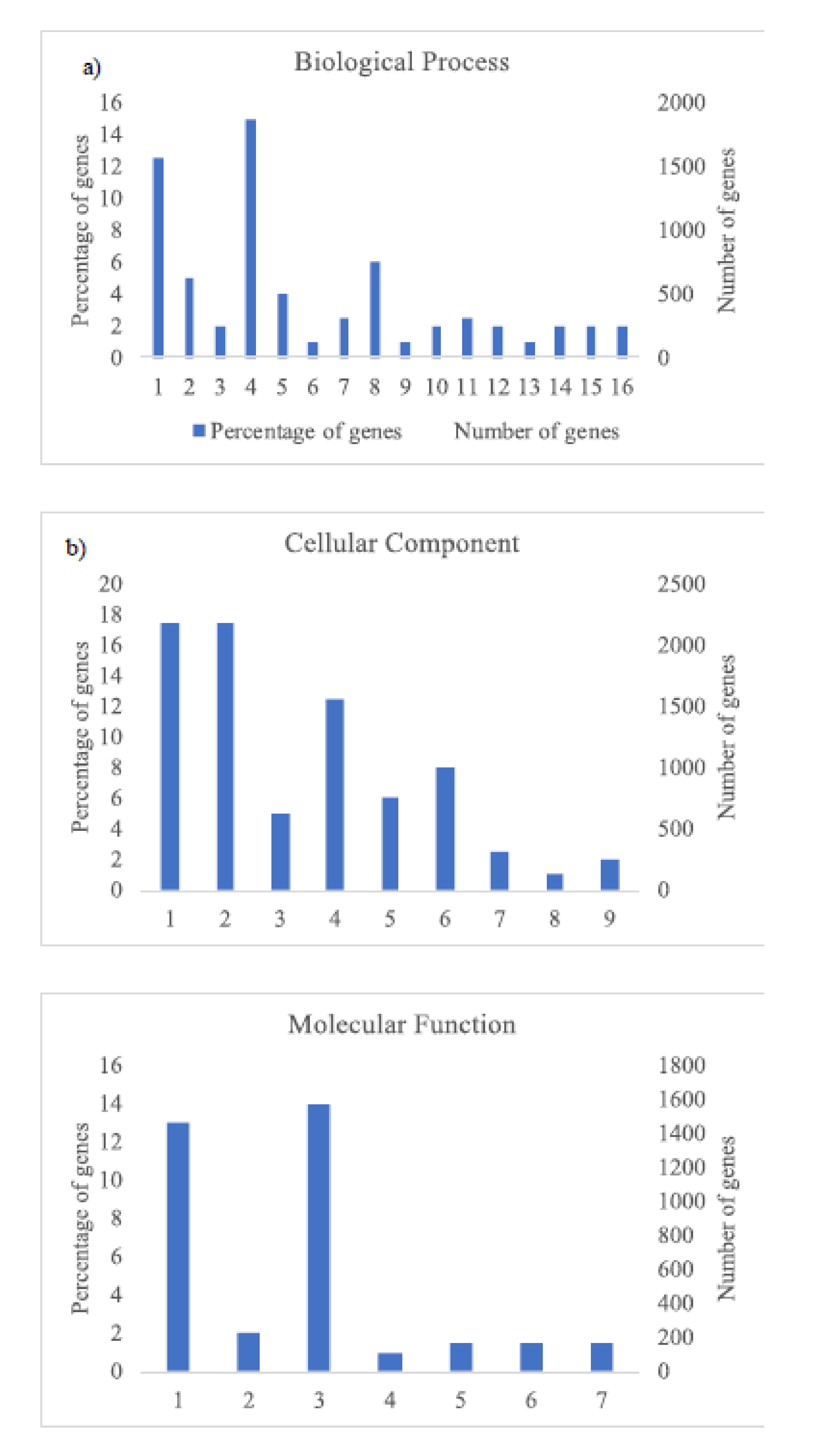
The GO analysis (Figure 3 and Figure 4) shows the most abundant genes grouped in three categories: Cellular components, molecular function, and biological process. Common to both years, the most abundant genes corresponded to “cell parts”, “organelle” and “cell” subcategories within “cellular components” category, “catalytic activity” and “binding” subcategories within “molecular function” category, and “metabolic process” and “cellular process” within “biological process” category and specific to 2018, “biological regulation” and “response to stimulus” within “biological process” category (Figure 4).
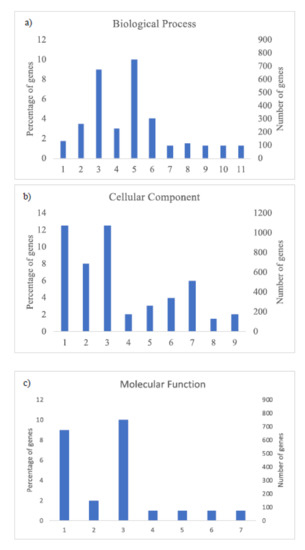
Figure 3.
Histogram of GO classifications of L. perenne pollen in samples of 2017. Results are summarized for the three main categories: (a) Biological process: 1. Localization, 2. Biological regulation, 3. Metabolic process, 4. Regulation of biological process, 5. Cellular process, 6. Response to stimulus, 7. Multi-organism process, 8. Cellular component organization or biogenesis, 9. Developmental process, 10. Multicellular organismal process, 11. Signaling; (b) cellular component: 1. Cell part, 2. Organelle, 3. Cell, 4. Protein-containing complex, 5. Organelle part, 6. Membrane part, 7. Membrane, 8. Membrane-enclosed lumen, 9. Extracellular region; and (c) molecular function: 1. Catalytic activity, 2. Transporter activity, 3. Binding, 4. Signal transducer activity, 5. Structural molecule activity, 6. Molecular function regulator, 7. Transcription regulator activity.
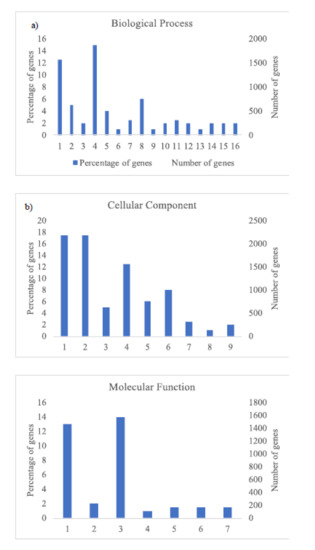
Figure 4.
Histogram of GO classifications of L. perenne pollen in samples of 2018. Results are summarized for the three main categories: (a) Biological process: 1. Metabolic process, 2. Biological regulation, 3. Multi-organism process, 4. Cellular process, 5. Regulation of biological process, 6. Negative regulation of biological process, 7. Localization, 8. Response to stimulus, 9. Immune system process, 10. Signaling, 11. Cellular component organization or biogenesis, 12. Developmental process, 13. Positive regulation of biological process, 14. Multicellular organismal process, 15. Reproductive process, 16. Reproduction; (b) cellular component: 1. Cell, 2. Cell part, 3. Organelle part, 4. organelle, 5. Membrane part, 6. Membrane, 7. Protein-containing complex, 8. Membrane-enclosed lumen, 9. Extracellular region; and (c) molecular function: 1. Catalytic activity, 2. Transporter activity, 3. Binding, 4. Molecular transducer activity, 5. Molecular function regulator, 6. Structural molecule activity, 7. Transcription regulator activity.
Among the upregulated genes in Madrid samples, ten showed isoforms in which overexpression was different depending on the year (Table 3); the number of isoforms from the six genes overexpressed in both years (HSP, glycoside hydrolase, Leucin rich repeat, EF hand family, pollen allergy, and coifilin), only in 2017 (Pectinesterase and serpin) and only in 2018 (lipid transfer protein and thaumatin) are indicated in the table. To make this table, the classification of functions proposed by the Pfam database (Supplementary Materials) has been taken into account. Only the genes overexpressed in Madrid have been taken into account since the greater allergenic capacity of this pollen with respect to that of Ciudad Real has already been shown, as well as its relationship with the atmospheric pollution. Therefore, the genes overexpressed in Madrid are candidates to be responsible for this greater allergenic capacity.

Table 3.
Protein encoded by the overexpressed genes in Madrid pollen samples.
3. Discussion
Nowadays, allergic diseases have become a pandemic health problem. Among them, pollen allergies are considered the most important [21]. Some studies showed that most of the patients sensitized to pollen allergens have perennial allergic rhinitis [21,22]. Moreover, it is demonstrated that these diseases appear to be more prevalent in industrialized countries, and the incidence seems to be higher in polluted areas, especially areas with heavy traffic [4,12,23].
Some studies have shown the existence of an in situ allergic response in patients with negative skin prick test (SPT) results and undetectable IgE in the serum [24]. This clinical entity, known as local allergic rhinitis (LAR) [25], is considered a new phenotype of allergic rhinitis (AR) that must be differentiated from nonallergic rhinitis [26,27]. This misunderstanding could be related to several facts: (i) In most cases, pollen to which patients are exposed is not the same as that used in skin prick tests; (ii) the number of allergens involved in the allergic processes may be greater than what has been described so far; and (iii) probably some of the allergens are only expressed upon specific physiological conditions of plants and, among which is the degree of atmospheric pollution [12].
The International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature Sub-committee (http://www.allergen.org/search.php?allergensource=Lolium+perenne) establishes six allergens in Lolium perenne (Lol p 1, Lol p2, Lol p 3, Lol p 4, Lol p 5, and Lol p 11). The first three are expansins, proteins specialized in pollination, with its role being to weaken the cell wall during the development of the pollen tube.
The study of differential expression by RNAseq for two consecutive years in the L. perenne pollen, in two cities with different levels of atmospheric pollution intends to identify new allergens of L. perenne in order to improve the immunogenic therapy, making it more effective. At the same time, determining allergens related to higher air pollution levels could shed some light to explain why many patients with a negative skin test, still show local signs of allergy.
Genes encoded for L.perenne pollen allergen 1 (Lol p 1), were overexpressed in Madrid samples, but they were by no means the most overexpressed. The most overexpressed genes in the Madrid pollen in the two years studied were heat shock proteins (HSP), specifically with a molecular weight of 20, 70, and 90 (Table 2). Most of them are from the chaperones family, and their mission is to refold damaged proteins after stressful situations [28]. They were first described in relation to heat stress situations, but they have been reported in many other stress situations. Some of these proteins have been described as allergy-causing agents in fungi, mites, chestnut (Cas s 9 is an HSP20), and hazelnut pollen (Cor a 10 is an HSP70) [29]. However, to date, the allergenicity of L. perenne pollen has not been related to HSP.
Other highly overexpressed genes in both years of study were those related to glycoside hydrolases (Table 2) (EC 3.2.1.), a widespread group of enzymes that hydrolyze the glycosidic bond between two or more carbohydrates, with a group of 100 different families according to the sequence similarity [30,31,32]. Family 17 showed the highest levels of expression in both years. Glycoside hydrolase family 17 includes enzymes with several activities such as endo-1,3-beta-glucosidase (EC3.2.1.39), lichenase (EC 3.2.1.73), or exo-1,3-glucanase (EC 3.2.1.58). Currently, these enzymes have only been found in plants and in fungi. Some glucanases from plants have been described as allergens, i.e., in Hevea brasiliensis latex [32], olive pollen [33], and plant foods [34]. However, there are no bibliographic references to these enzymes as putative allergens in L. perenne.
Leucine rich repeats (LRR) genes have also been detected to be overexpressed in pollen from Madrid in the two years studied. LRR are repeated sequences present in a number of proteins with diverse functions and cellular locations. These repeated sequences are usually involved in protein-protein interactions. LRR domains are composed of beta-alpha units that form curved horseshoe structures with a parallel beta sheet on the concave side and mostly helical elements on the convex side. LRR domains are often flanked by cysteine rich domains [35,36]. Nowadays, an LRR-containing protein from wheat was found by screening a phage display wheat cDNA library with wheat allergic patients’ IgE [37], but there is no bibliographic reference on L. perenne.
Genes of proteins with EF-HAND motifs were also overexpressed in the Madrid pollen in the two years studied. The EF-hand-containing proteins actively bind to Ca2+ and chelate the cytosolic calcium to regulate calcium homeostasis [38]. The major EF-hand containing proteins are calcium dependent protein kinases (CDPKs/CPKs), calcineurin B-like (CBL), calmodulin-like proteins (CMLs), and calmodulins (CaMs). Allergens of this type have been described in the pollen of many different plants as Alnus glutinosa (Aln g 4), Brassica napus (Bra n polcalcin), Chenopodium album (Che a 3), Olea europaea (Ole e 3 and Ole e 8), etc. However, there is no reference to L. perenne.
We have found that some genes overexpressed in Madrid in the first or second year of study, but not in both. During the first year, genes that code for pectinesterases (seven isoforms) were overexpressed. These enzymes play an important role in the cell wall metabolism during fruit ripening [39]. Sal k 1 from the Salsola kali pollen was shown to be a major allergen [40].
During the second year of study, seven genes that code for isoenzymes of lipid transfer proteins (LTPs) and six isoenzymes of thaumatin were overexpressed in Madrid. Both are described as pathogenesis-related (PR) proteins and have a reputation for their allergenic capacity. LPTs are from family 14 (PR 14) and thaumatins belong to family 5 (PR 5). To date, the International Union of Immunological Societies Allergen Nomenclature Subcommittee has reported 39 allergenic LTPs from vegetables (n = 7), pollen of trees and weeds (n = 9), fruits (n = 18), nuts and seeds (n = 4), as well as latex (n = 1) [41]. Thaumatin-like proteins (TLPs) have been known for years as the main allergens in some fruits and pollen, such as allergen 3 from Cupressus arizonica (Cup a 3) [20]. There is no report relating the allergenicity of L. perenne pollen neither for LPTs nor for TLPs.
4. Material and Methods
4.1. Pollen Used in the Study
Pollen from L. perenne plants growing in natural conditions in Ciudad Real and Madrid cities was used for the experiments. Pollen was collected from plants in the maximum pollen production period (mid of May 2017 and 2018). Three populations of L. perenne separated between 50 and 100 m were selected. Plants of each population were harvested and each one constituted a replicate. Pollen was recollected by the company Iberpolen S.L. (Alcala la Real, Jaen, Spain).
4.2. RNA Library Assembly
Before the RNA library assembly, ribosomal RNA was removed. This was performed with the Ribo-Zero rRNA kit removal kit. The TruSeq Stranded Total RNA library Prep kit was used to generate the libraries of RNA. First of all, 2 μg of total RNA (RIN > 9) libraries, were sequenced using a HiSeq2500 instrument (Illumina Inc, San Diego, CA, USA). Sequenced readings were paired-end with a length of 101 bp reading performed in six samples (three from Madrid and three from Ciudad Real). The estimated coverage was around 59 million reads per sample (one lane). Library generation and RNA sequencing was done at Sistemas Genómicos S.L. (Valencia, Spain) following the manufacturer’s instructions.
4.3. RNA Transcriptomics Analysis
The FastQC v0.11.4 tool was used to check the quality control of the raw data. Then, the raw paired-end reads were mapped against the “Lolium perenne” ASM173568v1 genome provided by the NCBI database using the Tophat2 2.1.0 algorithm [42]. Insufficient quality reads (phred score < 5) were eliminated using Samtools 1.2 [43] and Picard Tools 2.12.1. Then, the GC distribution (i.e., the proportion of guanine and cytosine bp along the reads) was assessed, this should have a desired distribution between 40–60%. Moreover, to confirm that our sequencing contained a small proportion of duplicates, the distribution of duplicates (quality of sequencing indicator) were evaluated. Expression levels were calculated using the HTSeq [44]. This method employs unique reads for the estimation of gene expression and filters the multi-mapped reads. Differential expression analysis between conditions was assessed using DESeq2 [45]. Finally, we selected differentially expressed genes with a p-value adjusted by FDR < 0.05 and a fold change of at least 1.5 [46]. The DEG analysis between pollen from Madrid and pollen from Ciudad Real was done by using statistical packages designed by Python and R. using the DESeq2 algorithm [45]. By applying a differential negative binomial distribution for the statistics significance [44], we identified the genes that were differentially expressed. We considered as differently expressed genes those with a FC value below −1.5 or higher than 1.5 and with a p-value (Padj) corrected by FDR ≤ 0.05 to avoid the identification of false positives across the differential expression data.
4.4. Functional Enrichment Analysis
The gene category enrichment analysis was performed by comparing the differentially expressed genes to the Uniprot database by using Blastx and setting an e-value of 0.01 and a minimum of 40% of the protein length/transcript ratio. With the obtained terms, an over representation test was performed using an in-house R Script developed at Sistemas Genómicos (Valencia, Spain). The graphical plotting of DEGs distribution within GO categories was performed at http://wego.genomics.org.cn/.
5. Conclusions
In conclusion, the results obtained in this work can be very useful, since we have described genes that code for some overexpressed proteins in conditions of higher air pollution with potential allergenic capacity for the first time in L. perenne. These proteins have to be synthesized by heterologous cloning before their allergenicity can be checked with skin tests. In the case of positive responses, our contribution to the knowledge of allergens of L. perenne and their relationship with atmospheric pollution would be confirmed. On the other hand, this would contribute to the development of much more effective vaccines, probably solving the problem of an allergic response in patients with a negative skin prick test (SPT).
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/11/1507/s1. Differential expression results obtained in the RNAseq analyses in 2017 and 2018. The description obtained in Gene Ontology (GO), KEGG Orthology (KO), Pfam, and Clusters of Orthologous Groups (COG) of each of the genes is also included.
Author Contributions
All authors are integrated into the research project PI15/00. The authors of the work come from different fields of science. F.F.B. and T.A. are allergists and researchers specializing in the relationship between air pollution and the incidence of allergies caused by pollen. In the design of the work, they have contributed to determine the time of pollen collection in collaboration with the company Iberpolen S.L., in addition to providing clinical descriptions derived from their experience in the consultations. J.A.L., E.G.-A., and F.J.G.-M., researchers specializing in plant physiology, interpreted the results obtained from RNAseq and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Economía y Competitividad, Instituto de Salud Carlos III of Spanish Government PI15/00, and co-founded by Fondo Europeo de Desarrollo Regional—FEDER for the Thematic Networks and Co-operative Research Centres: ARADyAL (RD16/0006/0028).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sinha, M.; Singh, R.P.; Kushwaha, G.S.; Iqbal, N.; Singh, A.; Kaushik, S.; Kaur, P.; Sharma, S.; Singh, T.P. Current Overview of Allergens of Plant Pathogenesis Related Protein Families. Sci. World J. 2014, 2014, 543195. [Google Scholar] [CrossRef]
- D´Amato, G.; Cecchi, L.; Bonini, S.; Nunes, C.; Annesi-Maesano, I.; Behrendt, H.; Liccardi, G.; Popov, T.; Cauwenberge, P. Allergenic pollen and pollen allergy in Europe. Allergy 2007, 62, 76–990. [Google Scholar] [CrossRef]
- Pawankar, R.; Canonica, G.W.; Holgate, S.T.; Lockey, R.F.; Blaiss, M. The WAO White Book on Allergy; Wisconsin World Allergy Organization: Milwaukee, WI, USA, 2013. [Google Scholar]
- Reinmuth-Selzle, K.; Kampf, C.J.; Lucas, K.; Lang-Yona, N.; Fröhlich-Nowoisky, J.; Shiraiwa, M.; Lakey, P.S.J.; Lai, S.; Liu, F.; Kunert, A.T.; et al. Air Pollution and Climate Change Effects on Allergies in the Anthropocene: Abundance, Interaction, and Modification of Allergens and Adjuvants. Environ. Sci. Technol. 2017, 51, 4119–4141. [Google Scholar] [CrossRef] [PubMed]
- Alfaya Arias, T.; Feo Brito, F.; García Rodríguez, C.; Pineda, F.; Lucas, J.A.; Gutierrez Mañero, F.J.; Guerra, F. Lolium perenne pollen from a polluted city shows high allergenic potency and increased associated enterobacteriaceae counts. J. Investig. Allergol. Clin. Immunol. 2014, 24, 132–134. [Google Scholar]
- Armentia, A.; Lombardero, M.; Callejo, A.; Barber, D.; Gil, F.J.M.; Martín-Santos, J.M.; Vega, J.M.; Arranz, M.L. Is Lolium pollen from an urban environment more allergenic than rural pollen? Allergol. Immunopathol. 2002, 30, 218–224. [Google Scholar] [CrossRef]
- Feo Brito, F.; Mur Gimeno, P.; Martínez, C.; Tobías, A.; Suárez, L.; Guerra, F.; Borja, J.M.; Alonso, A.M. Air pollution and seasonal asthma during the pollen season. A cohort study in Puertollano and Ciudad Real (Spain). Allergy 2007, 62, 1152–1157. [Google Scholar] [CrossRef]
- Frank, U.; Ernst, D. Effects of NO2 and Ozone on Pollen Allergenicity. Front. Plant Sci. 2016, 7, 836. [Google Scholar] [CrossRef]
- Ghiani, A.; Aina, R.; Asero, R.; Bellotto, E.; Citterio, S. Ragweed pollen collected along high-traffic roads shows a higher allergenicity than pollen sampled in vegetated areas. Allergy 2012, 67, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Mur Gimeno, P.; Feo Brito, F.; Martínez, C.; Tobías, A.; Suárez, L.; Guerra, F.; Galindo, P.A.; Gómez, E. Decompensation of pollen-induced asthma in two towns with different pollution levels in La Mancha, Spain. Clin. Exp. Allergy 2007, 37, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Sénéchal, H.; Visez, N.; Charpin, D.; Shahali, Y.; Peltre, G.; Biolley, J.P.; Lhuissier, F.; Couderc, R.; Yamada, O.; Malrat-Domenge, A.; et al. A Review of the Effects of Major Atmospheric Pollutants on Pollen Grains, Pollen Content, and Allergenicity. Sci. World J. 2015, 2015, 940243. [Google Scholar] [CrossRef]
- Lucas, J.A.; Gutierrez Albanchez, E.; Alfaya, T.; Feo-Brito, F.; Gutierrez Mañero, F.J. Oxidative stress in ryegrass growing under different air pollution levels and its likely effects on pollen allergenicity. Plant Physiol. Biochem. 2019, 135, 331–340. [Google Scholar] [CrossRef]
- Emberlin, J. Grass, tree, and weed pollen. In Allergy and Allergic Diseases; Kay, A.B., Kapan, A.P., Bousquet, J., Holt, P.G., Eds.; Wiley-Blackwell: Oxford, UK, 2009; pp. 942–962. [Google Scholar]
- Hrabina, M.Ã.; Peltre, G.; Van Ree, R.; Moingeon, P.Ã. Grass pollen allergens. Clin. Exp. Allergy Rev. 2008, 3, 7–11. [Google Scholar] [CrossRef]
- Taketomi, E.A.; Sopelete, M.N.C.; de Sousa Moreira, P.F.; de Assis Machado Vieira, F. Pollen allergic disease: Pollens and its major allergens. Braz. J. Otorhinolaryngol. 2006, 72, 562–567. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, J.H.; Park, K.H.; Kim, K.R.; Han, M.J.; Choe, H.; Oh, J.W.; Hong, C.S. A six-year study on the changes in airborne pollen counts and skin positivity rates in Korea: 2008–2013. Yonsei Med. J. 2016, 57, 714–717. [Google Scholar] [CrossRef]
- Marsh, D.G. Allergens and the genetics of allergy. In The Antigens; Sela, M., Ed.; Academic Press: New York, NY, USA, 1975; Volume 3, pp. 271–350. [Google Scholar]
- Subiza, J. Gramíneas: Aerobiología y polinosis en España. Rev. Esp. Alergol. Inmunol. Clín. 2003, 18, 7–23. [Google Scholar]
- Kusano, H.; Li, H.; Minami, H.; Kato, Y.; Tabata, H.; Yazaki, K. Evolutionary Developments in Plant Specialized Metabolism, Exemplified by Two Transferase Families. Front. Plant Sci. 2019, 10, 683. [Google Scholar] [CrossRef]
- Breiteneder, H. Thaumatin-like proteins—A new family of pollen and fruit allergens. Allergy 2004, 59, 479–481. [Google Scholar] [CrossRef]
- Chen, M.; Xu, J.; Devis, D.; Shi, J.; Ren, K.; Searle, I.; Zhang, D. Origin and Functional Prediction of Pollen Allergens in Plants. Plant Physiol. 2016, 172, 341–357. [Google Scholar] [CrossRef]
- Sedghy, F.; Varasteh, A.R.; Sankian, M.; Moghadam, M. Interaction Between Air Pollutants and Pollen Grains: The Role on the Rising Trend in Allergy. Rep. Biochem. Mol. Biol. 2018, 6, 219–224. [Google Scholar]
- García-Gallardo, M.V.; Algorta, J.; Longo, N.; Espinel, S.; Aragones, A.; Lombardero, M.; Bernaola, G.; Jauregui, I.; Aranzabal, A.; Albizu, M.V.; et al. Evaluation of the effect of pollution and fungal disease on Pinus radiata pollen allergenicity. Int. Arch. Allergy Immunol. 2013, 160, 241–250. [Google Scholar] [CrossRef]
- Blanca-Lopez, N.; Campo, P.; Salas, M.; García Rodríguez, C.; Palomares, F.; Blanca, M.; Canto, G.; Feo Brito, F.; Rondon, C. Seasonal Local Allergic Rhinitis in Areas With High Concentrations of Grass Pollen. J. Investig. Allergol. Clin. Immunol. 2016, 26, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Rondón, C.; Doña, I.; López, S.; Campo, P.; Romero, J.J.; Torres, M.J.; Mayorga, C.; Blanca, M. Seasonal idiopathic rhinitis with local inflammatory response and specific IgE in absence of systemic response. Allergy 2008, 63, 1352–1358. [Google Scholar] [CrossRef]
- Rondon, C.; Fernandez, F.; Canto, G.; Blanca, M. Local allergic rhinitis: Concept, clinical manifestations, and diagnostic. J. Investig. Allergol. Clin. Immunol. 2010, 20, 364–371. [Google Scholar]
- Rondón, C.; Campo, P.; Togias, A.; Fokkens, W.J.; Durham, S.R.; Powe, D.G.; Mullol, J.; Blanca, M. Local allergic rhinitis: Concept, pathophysiology, and management. J. Allergy Clin. Immunol. 2012, 129, 1460–1467. [Google Scholar]
- Binder, R.J. Functions of heat shock proteins in pathways of the innate and adaptive immune system. J. Immunol. 2014, 193, 5765–5771. [Google Scholar] [CrossRef]
- Gruehn, S.; Suphioglu, C.; O’Hehir, R.E.; Volkmann, D. Molecular cloning and characterization of hazel pollen protein (70 kD) as a luminal binding protein (BiP): A novel cross-reactive plant allergen. Int. Arch. Allergy Immunol. 2003, 131, 91–100. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G.J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000, 124, 1515–1519. [Google Scholar] [CrossRef]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef]
- Yagami, T.; Osuna, H.; Kouno, M.; Haishima, Y.; Nakamura, A.; Ikezawa, Z. Significance of carbohydrate epitopes in a latex allergen with beta-1,3-glucanase activity. Int. Arch. Allergy Immunol. 2002, 129, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Palomares, O.; Villalba, M.; Quiralte, J.; Polo, F.; Rodriguez, R. 1,3-beta-glucanases as candidates in latex-pollen-vegetable food cross-reactivity. Clin. Exp. Allergy 2005, 35, 345–351. [Google Scholar] [CrossRef]
- Wagner, S.; Radauer, C.; Hafner, C.; Fuchs, H.; Jensen-Jarolim, E.; Wuthrich, B.; Scheiner, O.; Breiteneder, H. Characterization of cross-reactive bell pepper allergens involved in the latex-fruit syndrome. Clin. Exp. Allergy 2004, 34, 1739–1746. [Google Scholar] [CrossRef] [PubMed]
- Enkhbayar, P.; Kamiya, M.; Osaki, M.; Matsumoto, T.; Matsushima, N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins 2004, 54, 394–403. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Weichel, M.; Vergoossen, N.J.; Bonomi, S.; Scibilia, J.; Ortolani, C.; Ballmer-Weber, B.K.; Pastorello, E.A.; Crameri, R. Screening the allergenic repertoires of wheat and maize with sera from double-blind, placebo-controlled food challenge positive patients. Allergy 2006, 61, 128–135. [Google Scholar] [CrossRef]
- Mohanta, T.; Yadav, D.; Khan, A.; Hashem, A.; Abd Allah, E.; Al-Harrasi, A. Molecular Players of EF-hand Containing Calcium Signaling Event in Plants. Int. J. Mol. Sci. 2019, 20, 1476. [Google Scholar] [CrossRef]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef]
- Barderas, R.; Garcia-Selles, J.; Salamanca, G.; Colas, C.; Barber, D.; Rodriguez, R.; Villalba, M. A pectin methylesterase as an allergenic marker for the sensitization to Russian thistle (Salsola kali) pollen. Clin. Exp. Allergy 2007, 37, 1111–1119. [Google Scholar] [CrossRef]
- Egger, M.; Hauser, M.; Mari, A.; Ferreira, F.; Gadermaier, G. The role of lipid transfer proteins in allergic diseases. Curr. Allergy Asthma Rep. 2010, 10, 326–335. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).