Abstract
Benzobicyclon has shown varying results in controlling weedy rice, including those with imidazolinone (IMI) resistance. Tolerance to benzobicyclon in cultivated japonica rice, but not indica or aus-like cultivars, is conferred by a fully functional HPPD Inhibitor Sensitive 1 (HIS1) gene. Herein, a diagnostic Kompetitive Allele Specific PCR (KASP) assay was developed to predict the HIS1 genotype of weedy rice plants from 37 accessions and correlated to their response to benzobicyclon in the field. Two-thirds of the 693 weedy rice plants screened were tolerant to benzobicyclon (371 g ai ha−1, SC formulation) at 30 days after treatment (DAT). Thirty-four percent of plants were homozygous for the HIS1 allele and 98% of these plants exhibited field tolerance. However, the his1 genotype did not always correlate with field data. Only 52% of his1 plants were considered sensitive, indicating that the single nucleotide polymorphisms (SNPs) chosen in the KASP assay are not a reliable tool in predicting his1 homozygous plants. In an additional experiment, 86% of the 344 plants with at least one copy of the ALSS653N trait harbored a HIS1 allele, suggesting fields infested with IMI herbicide-resistant weedy rice are unlikely to be controlled with benzobicyclon.
1. Introduction
In rice (Oryza sativa L.) fields worldwide, weedy rice is a troublesome weed that can cause economic losses by reducing grain yield and/or grain quality [1,2]. Season-long interference of weedy rice at 2 or 40 plants m−2 was found to reduce the grain yield of different cultivars by 7 to 19% and 61 to 87%, respectively [3,4]. Taxonomically, weedy rice and cultivated rice are the same species, and thus selective chemical control of weedy rice in cultivated rice is extremely difficult. Cultural practices such as crop rotation, stale seedbed, water seedings, or transplanting instead of direct-seeding are some of the methods used around the world to deter weedy rice infestations [5]. In the 2000s, non-transgenic IMI-resistant (ClearfieldTM technology, BASF) rice became commercially available in the midsouthern United States and served as an effective tool for controlling many weeds of rice, including weedy rice. IMI-resistant cultivars have a mutation in the acetolactate synthase (ALS) gene. The original Clearfield lines contained a mutated ALSG654E gene, but were quickly replaced by cultivars harboring the ALSS653N trait because of their superior yield and improved tolerance to IMIs [2,6,7]. Unfortunately, owing to the natural gene flow between IMI-resistant rice and weedy rice in conjunction with the overreliance of IMIs, IMI-resistant weedy rice accessions quickly appeared and are now prevalent throughout the rice growing regions in the midsouthern United States [8].
The pro-herbicide benzobicyclon is a 4-hydroxyphenylpyruvate dioxygenase (HPPD)-inhibitor under registration in the midsouthern United States for control of several annual grasses, sedges, broadleaves, and aquatic weeds in flooded rice [9,10]. Rice sensitivity to benzobicyclon is cultivar-specific, but at broad-scale, tolerance diverges among rice subspecies. The two major cultivated rice subspecies worldwide are japonica and indica, where japonica cultivars are typically tolerant, while indica cultivars are highly sensitive [10,11]. In japonica cultivars, tolerance to benzobicyclon is conferred by a fully-functional HIS1 gene found on chromosome two in the rice genome. HIS1 belongs to the Fe(II)/2OG-dependent oxygenase family, similar to the HPPD enzyme, and was found to hydroxylate benzobicyclon hydrolysate and not benzobicyclon into a less phytotoxic compound [12]. The his1 allele in indica cultivars contains a 28 bp deletion loss-of-function mutation that results in an early stop codon and encodes for a severely truncated protein [12]. Additionally, the HIS1 gene in aus rice cultivars such as Kasalath [13] and Purple Marker [10] do not contain a 28 bp deletion, yet aus cultivars appear to be sensitive to benzobicyclon and will hereafter be referred to as HIS1 *. In comparison with the HIS1 gene in japonica cultivars, several mutations can be found near the 3′ end of the HIS1 * gene in aus cultivars and may alter protein function, but this has not been proven.
Weedy rice in the midsouthern United States is morphologically and phenotypically diverse with two suggested groups based on seed characteristics: strawhull (awnless) and blackhull (awned) [14]. Genetic analysis by Shivrain et al. [15] indicated weedy rice arose from hybridizations events between O. rufipogon, O. sativa indica, and aus, while Reagon et al. did not find unambiguous evidence for genetic contribution from O. rufipogon [16]. However, significant admixture has occurred between weedy rice and the popular japonica cultivars grown in the midsouth (e.g., Clearfield-resistant weedy rice). Interestingly, in benzobicyclon-containing research plots in Arkansas, benzobicyclon was serendipitously found to have significant activity on weedy rice [17]. This prompted Young and co-authors [10] to access the sensitivity of weedy rice accessions collected across the midsouthern United States to benzobicyclon. The authors found the response of weedy rice plants to benzobicyclon at 371 g ha−1 (SC formulation) varied considerably within and among accessions. In addition, they found that, in the field, 30 out of the 100 accessions screened exhibited at least 80% control (~50 plants plot area−1).
In the current study, the benzobicyclon sensitivity of 37 weedy rice accessions used by Young et al. [10] was further dissected. Specifically, 14 to 24 weedy rice plants per accession were subjected to benzobicyclon in the field and later genotyped for the HIS1/his1 alleles and for the ALSS653N trait using KASP. KASP is a common quantitative PCR genotyping technique designed to identify allele specific SNPs [18]. The overall objective of this study was to develop diagnostic tools to predict the effectiveness of benzobicyclon in a growers’ weedy rice management program.
2. Results and Discussions
2.1. Sensitivity of Weedy Rice to Benzobicyclon
The percent control of 37 different weedy rice accessions (14 to 24 plants per accession) when treated with benzobicyclon (371 g ha−1) at 30 DAT averaged 28% (Table S1). Based on the response of all plants within an accession, 21, 8, and 8 of the accessions tested were classified (>90% of plants) as benzobicyclon tolerant, mixed, or susceptible, respectively. On a plant basis, 69% of the 693 plants screened were classified as benzobicyclon tolerant. Of the remaining 216 sensitive plants, 77% were killed, with the rest significantly injured (>50% injury) at 30 DAT. These results are consistent with Young and co-authors [10], who found benzobicyclon did not adequately (<80%) control 70 out of the 100 accessions screened and noted several accessions were a heterogenous mixture of sensitive and tolerant weedy rice plants.
2.2. HIS1 Genotype of Weedy Rice Plants
The 693 plants from the field assay were genotyped for the HIS1 gene using a KASP assay. The assay was designed to distinguish between the HIS1 allele found in japonica rice and the HIS1 */his1 alleles found in sensitive aus/indica cultivars. Although plants harboring the HIS1 * or his1 alleles have differing levels of sensitivity to benzobicyclon [13], both allelic forms in this assay are considered sensitive and cannot be differentiated. HIS1 */his1 will hereafter be referred to as his1. Sanger sequencing results agreed with the KASP assay calls, which highlights the reliability of our assay to differentiate between HIS1 and his1 alleles based on the chosen SNPs (Figure 1).
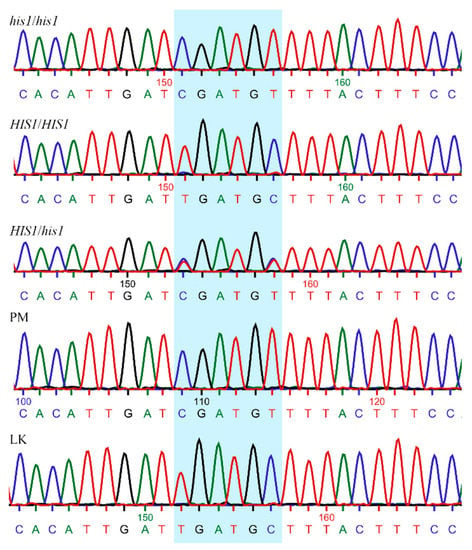
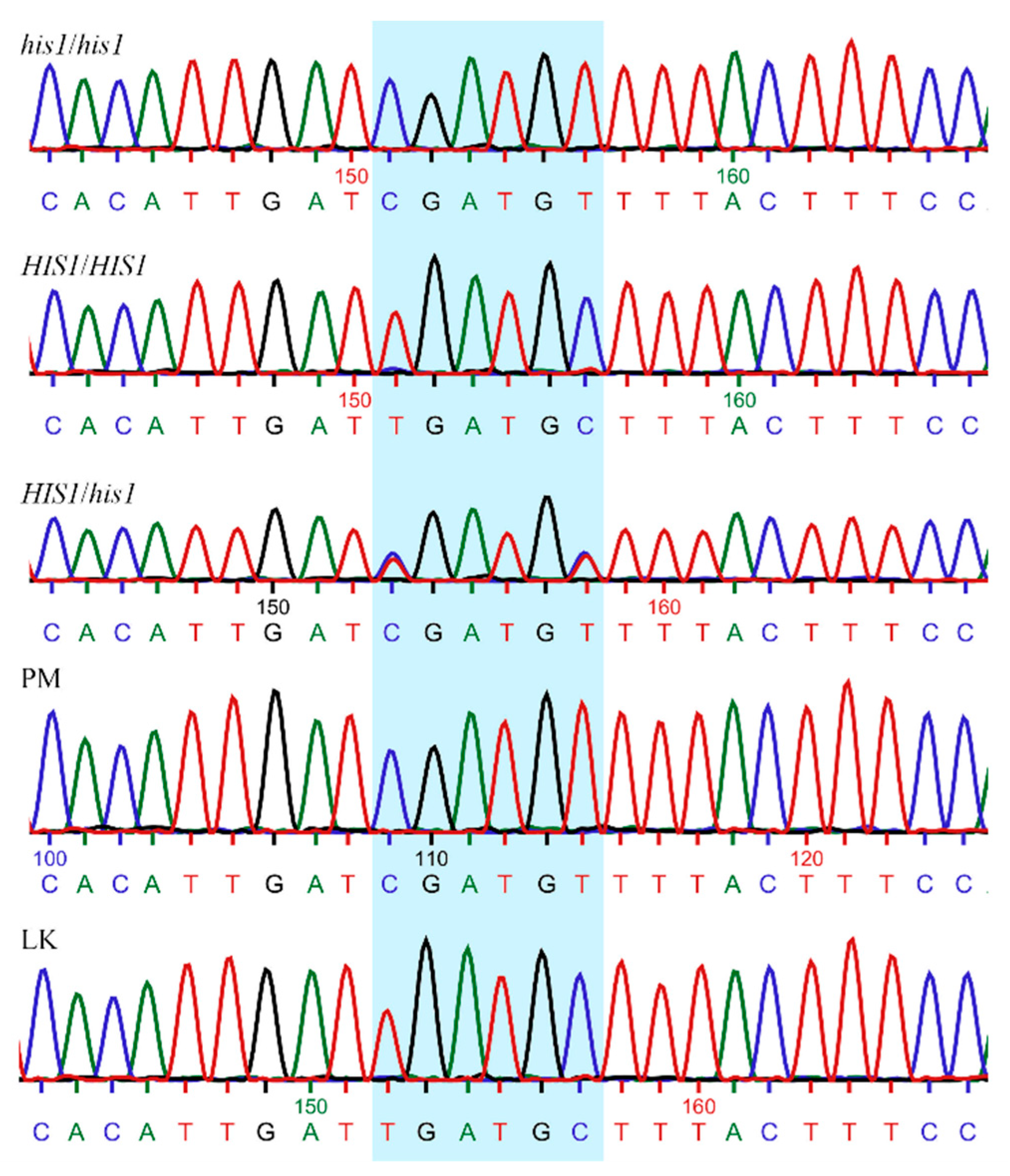
Figure 1.
Chromatograms of Sanger sequencing showing partial HIS1 sequence and the target used (highlighted) for genotyping the different weedy rice accessions. Rice varieties known for their susceptibility (PM) and tolerance to benzobicyclon (LaKast) were used as reference. his1/his1: homozygous susceptible; HIS1/HIS1: homozygous resistant; HIS1/his1: heterozygous; PM: Purple Marker; LK: LaKast.
To determine a typical response of weedy rice plants within an accession, we broadly classified accessions based on their HIS1 allele frequency; where an accession was considered tolerant if ≥90% of the alleles were HIS1, mixed if 10% < HIS1 < 90%, or sensitive if HIS1 ≤ 10%. Out of the 37 accessions screened, 7, 15, and 15 were predicted to be a tolerant, mixed, or sensitive population, respectively (Table 1 and Table S1). All seven accessions classified as tolerant in the KASP assay were classified as tolerant in the field experiment and, furthermore, on a plant level, 98% of all homozygous HIS1 plants were biologically tolerant to benzobicyclon. However, the genotypic classification of an accession was not always analogous with the field results. In fact, only 18 out of the 37 accessions tested had similar classifications. These results indicate that the SNPs chosen in the KASP assay are not a reliable tool in predicting his1 homozygous plants’ sensitivity to benzobicyclon.

Table 1.
Biological and genotyping designation of the 37 weedy rice accessions used in this study. Data correlate the sensitivity to benzobicyclon in weedy rice plants at field level, and the genotyping results of the HIS1 and the ALSS653N trait.
In the 19 accessions with contradicting classifications, the difference in two accessions classified as biologically sensitive, but genotyped as a HIS1/his1 mixed population (accessions 18 and 23), could possibly be explained by the high level of benzobicyclon sensitivity observed in heterozygous plants. In the remaining 17 accessions, the discrepancy can primarily be attributed to the erroneous classification of plants in the KASP assay as his1. Fourteen of the 19 misclassified accessions were considered tolerant in the field, but classified as a HIS1/his1 mixed population or sensitive (his1/his1). The other three accessions were predicted to be sensitive, but were a mixture of biologically tolerant and sensitive plants within each accession. These results can be further exemplified by comparing the plant genotype to its biological response to benzobicyclon across accessions. Not surprisingly, 98% of the 216 biologically sensitive plants were either homozygous or heterozygous for the his1 allele; however, an additional 180 plants predicted to be homozygous for the his1 allele were found to be tolerant to benzobicyclon (Figure 2).
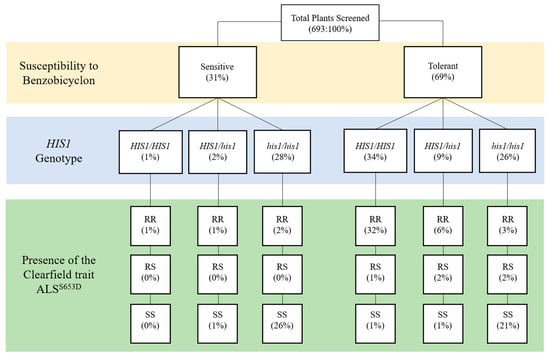
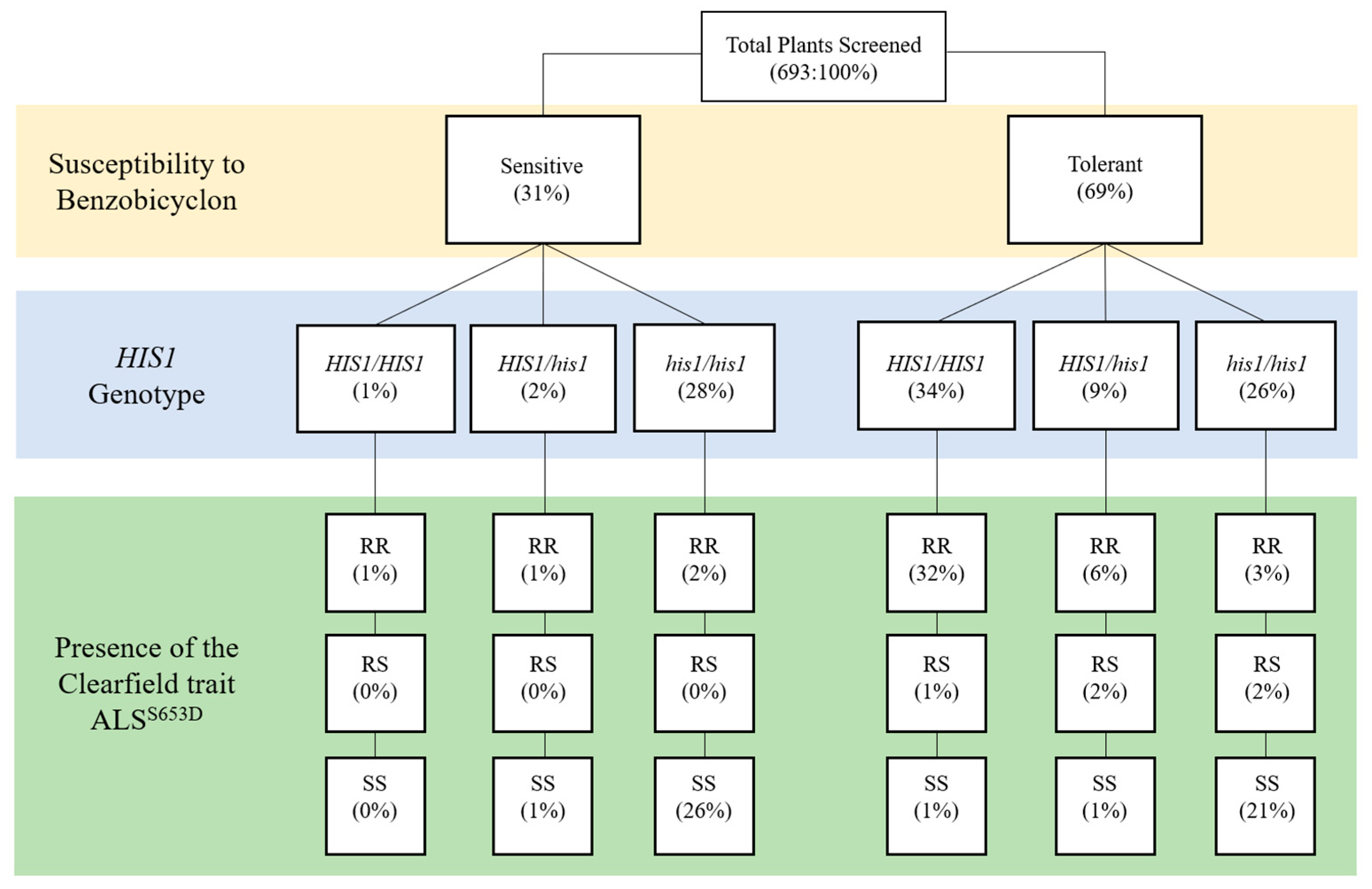
Figure 2.
Dendogram describing the relationship between benzobicyclon susceptibility and the results from the HIS1 and the Clearfield trait ALSS653N Kompetitive Allele Specific PCR (KASP) assays. HIS1/HIS1: homozygous resistant; HIS1/his1: heterozygous; his1/his1: homozygous susceptible; RR: homozygous resistant; RS: heterozygous; SS: homozygous susceptible. Values in parenthesis are the percentage of plants with that specific genotype.
2.3. Correlation between HIS1 and the ALSS653N Clearfield Trait
In Arkansas, Clearfield-resistant japonica rice with the ALSS653N mutation has high market penetration [19]; however, owing to gene flow, IMI-resistant weedy rice is now a widespread issue in many grower fields [8]. Interestingly, the ALSS653N trait and the HIS1 loci are localized to the lower half of chromosome two in the rice genome. Here, we are not interested in the severity of IMI-resistant weedy rice, but what is the probability the ALSS653N and the HIS1 genes co-segregate in the likely event of gene flow between weedy rice and the japonica cultivars grown in Arkansas. Thus, the 693 plants genotyped for HIS1 were also genotyped for the presence or absence of the ALSS653N trait using a KASP assay [7]. Figure 2 classifies plants by their biological response to benzobicyclon and their subsequent genotype for HIS1 and ALSS653N. As expected, IMI-resistant weedy rice was a common genotype in 22 out of the 37 accessions (Table 1) and, in fact, 344 out of 693 plants screened contained at least one copy of the ALSS653N (homozygous resistant (RR) and heterozygous (RS)) trait (Figure 2). The probability that the ALSS653N trait and HIS1 co-segregate was found to be high (ꭓ2 = 456, p < 0.0001). In the 344 plants with at least one copy of the ALSS653N trait, 86% also harbored a HIS1 allele. These results suggest that IMI resistance in weedy rice obtained via gene flow from the commonly grown japonica cultivars in the midsouth United States could be used as an indicator for predicting the tolerance of weedy rice to benzobicyclon.
In this study, a KASP assay was designed to have a rapid diagnostic tool to predict the sensitivity of weedy rice accessions to benzobicyclon. The assay was designed to genotype plants for the presence or absence of a fully functional HIS1 allele normally found in japonica rice, but not in sensitive indica or aus cultivars. Unfortunately, the predicted HIS1 genotype from the KASP assay did not always match the field data. For example, the KASP assay predicted 374 benzobicyclon sensitive homozygous his1 plants, but only 52% of these were killed or significantly injured in the field. It is possible the predicted susceptible, but actually tolerant weedy rice might carry a unique resistant mechanism (e.g., point mutations in HPPD target gene or alternative detoxification route) or, alternatively, the heterogenous genetic makeup of weedy rice could play a role in the discrepancy with our KASP assay. Weedy rice in the midsouthern United States is believed to be derived from indica and aus subpopulations, but the genetic contribution from the wild rice progenitor, O. rufipogon, has contradicting reports [15,16,20]. The HIS1 genotype of O. rufipogon is unknown and it is plausible that O. rufipogon harbors a functional HIS1 gene that shares the SNPs used in our assay to identify benzobicyclon sensitive indica or aus his1 alleles. The susceptibility of O. rufipogon to benzobicyclon needs to be determined to further explore this theory. Regardless, our results indicate that a new assay designed around alternative SNPs is needed to better predict the sensitivity of weedy rice to benzobicyclon. Despite the above, our field results indicate benzobicyclon alone cannot be relied upon as a viable option for weedy rice control in the midsouthern United States, which further complements the findings of Young et al. [10]. Even more troublesome, a high correlation (86%) between the prevalence of the ALSS653N IMI-resistant trait and the HIS1 gene in weedy rice was found. These findings suggest that, in fields infested with IMI-resistant weedy rice, the probability benzobicyclon will effectively kill these plants is low. However, it should be noted that benzobicyclon had considerable activity on heterozygous HIS1 plants in accessions where the homozygous his1 plants were sensitive in the field. Previous research has shown benzobicyclon can significantly injure transplanted japonica × indica cultivars [11,21,22,23]. For example, Kim et al. [23] found three transplanted japonica × indica cultivars, on average, exhibited 50% foliar chlorosis at 14 DAT and the tiller number per hill was reduced by one-third after treatment with benzobicyclon at 350 g ha−1.
Further research is needed to determine the response of homozygous and heterozygous HIS1 plants at different growth stages in the presence and absence of crop competition, considering that previous research has shown weedy rice control can partially be a function of size at application [24,25]. Regardless, weedy rice control in non-transgenic cultivated rice will remain a difficult endeavor and an integrated weed management approach is required.
3. Materials and Methods
3.1. Plant Material
The 37 weedy rice accessions used in this study are a subsample of accessions used by Young et al. [10]. The accessions were originally collected in 2015 from rice fields predominantly in Arkansas, but also in Mississippi and Missouri. A detailed sampling methodology and collection sites (GPS coordinates) can be found therein.
3.2. Benzobicyclon Field Study
In the summer of 2019, a field experiment was conducted to investigate and correlate the response of weedy rice to benzobicyclon at the plant, accession, and genotypic levels. The experiment was conducted at the Milo J. Shult Agricultural Research and Extension Center in Fayetteville, AR. The previous year’s crop was soybean and the soil type was a leaf silt-loam soil (fine, mixed, active, thermic Typic albaquults) with a pH of 5.2 and 1.8% organic matter content. During the experiment, the maximum and minimum temperatures registered were 29.4 °C and 19.3 °C, respectively, with a mean temperature of 24.3 °C, and the average precipitation was 92.5 mm. The experiment was a one-factor (weedy rice accession) randomized complete block design. The entire experiment was contained in a single bay of 18 by 25 m with a 0.9 m alley separating three replications. In each block, an accession was hand planted in rows distanced 30 cm, and within a row, ten total hills (three seeds hill−1) were planted at 0.3 m spacings. Each accession was replicated and randomly located once within a block. The total number of plants tested for an accession ranged from 14 to 24 plants owing to unforeseen circumstances (poor germination, plants too small at application, or insufficient DNA yield after extraction). Non-treated plants were not included in this experiment, however, the japonica benzobicyclon-tolerant cultivar ‘LaKast’ and a benzobicyclon-sensitive aus cultivar ‘Purple Marker’ were included as internal tolerant and sensitive controls, respectively. Plots were fertilized and maintained weed-free using Arkansas extension recommendations for rice production [26].
Once plants reached the two-leaf stage, a hill was flagged and thinned to one plant, then at the four- to five-leaf growth stage, a 5 to 10 cm flood was established three days prior to benzobicyclon treatment and the flood was maintained for 30 DAT. At this time, plants with less than four to five leaves were pulled to remove the potential confounding effect of plant size. At application, benzobicyclon at 371 g ai ha−1 (RogueTM, 3.3 SC, Gowan Co, Yuma, AZ, USA) plus methylated seed oil (1% v/v) was applied across the entire test area with a CO2-pressurised backpack sprayer with a four-nozzle handheld boom equipped with 110015 AIXR nozzles calibrated to deliver 143 L ha−1 at 276 kPa. At 30 DAT, plant injury was recorded on a scale from 0 to 100%, where 0 equaled a healthy plant with no visible reduction in plant growth or injury (e.g., bleaching symptomology characteristic of HPPD-inhibitors) and 100% being plant death. The response of the tolerant (LaKast) and sensitive (Purple Marker) controls were used as baselines. An accession was classified as tolerant or susceptible if ≥90% of the plants exhibited the same response.
3.3. DNA Extraction
Plant tissue (top third section of newly emerged leaf) of weedy rice accessions was collected at the two-leaf stage, placed in individual Eppendorf tubes, and stored at −80 °C for subsequent DNA extraction. DNA was isolated from weedy rice tissue using the CTAB (cetyl trimethylammonium bromide) protocol modified for a 96-well plate [27]. Tissue was then placed in 1.2 mL tall microtubes and frozen tissue was finely ground with two metal beads (2.4 mm diameter) on a mixer mill (Retsch, MM400) at a frequency of 30 hz s−1 during 15 s. Then, 300 µL of CTAB (2% CTAB, 1.4 M NaCl, 20 mM EDTA (ethylenediamine tetraacetic acid), and 100 mM Tris-HCI, pH 8.0) extraction buffer was added to each sample. DNA quality and quantity were assessed spectrophotometrically (Nanodrop 2000c, Thermo Scientific, Waltham, MA, USA).
3.4. KASP Assay
KASP assays were designed to detect the Clearfield ALSS653N trait and HIS1 alleles. The ALSS653N KASP assay used in this study was previously created by Rosas et al. [7], and the protocol provided therein was followed. The HIS1 KASP assay was designed to differentiate between a fully functional HIS1 gene (e.g., HIS1 in japonica cultivars) from the HIS1 gene found in the benzobicyclon susceptible cultivars of indica (his1) and aus (HIS1 *). The dataset from the 3k Rice Genome Project (http://snp-seek.irri.org [28]) was used to identify SNPs shared between indica and aus cultivars, but not found in the HIS1 gene of japonica. SNPs localized in the second intron of HIS1 at the 607th and 612th nucleotides (GAT(T/C)GATG(C/T)TTTA; tolerant/susceptible) were chosen for assay development. The SNPs of interest were sent to LGC Genomics (Beverly, MA, USA) for in-house primer design.
KASP reactions comprised a 2× KASP Master Mix, KASP Assay mix (primers), and weedy rice DNA. The reactions were run on 96-well plates on a CFX Connect Real-Time System (Bio-Rad Laboratories Inc., Hercules, CA, USA) following the directions of the manufacturer. Thermal cycle was as follows: 1 cycle of activation at 94 °C for 15 min, 10 cycles consisting of a denaturation step at 94 °C for 20 s, annealing/elongation of 61 °C (decreasing 0.6 °C per cycle) for 1 min, and 26 cycles with a denaturation step of 94 °C for 20 s and 55 °C for 1 min. Plate read was performed after a cycle at 37 °C for 1 min. Allelic discrimination was automatically assigned by the CFX Maestro software based on the relative fluorescence units of FAM and HEX fluorophores obtained at the end of the run. For the ALSS653N trait assay, the cultivar LaKast was used in all reactions as a homozygous susceptible (SS) control. For the HIS1 assay, LaKast (HIS1/HIS1), Purple Marker (HIS1 */HIS1 *), and a synthetic heterozygous mixture of LaKast/Purple Marker (1:1 ratio) served as homozygous tolerant, homozygous susceptible, and heterozygous standards. A no-template control where water was adding instead of DNA was included in all runs.
3.5. HIS1 Gene Sequencing
To verify the results from the HIS1 KASP assay, the HIS1 gene from three randomly selected weedy rice samples from each class (homozygous tolerant, sensitive, and heterozygous) plus LaKast and Purple Marker were sequenced. The primers used to amplify a 300 bp amplicon centered around the SNPs of interest were the forward primer 5′ATGGCAAGAACTTCCAGATTC 3′ and the reverse primer 5′AGACAACAATCCTTGTTACTTTGTAAG 3′. Each 50 µL PCR reaction consisted of 1× Colorless GoTaq Flexi Buffer (Promega Corp., WI, USA), 1.5 mM MgCl2, 0.2 mM dNTP’s, 0.2 µM of each forward and reverse primer, 100 ng DNA, and 1.25 units GoTaq Hot Start Polymerase. PCR conditions were as follows: 94 °C 2 min, 35 cycles of 94 °C 30 s, 51 °C 30 s, 72 °C 1 min, and a final extension of 72 °C 5 min. An aliquot of the PCR product was run on a 1.5% w/v agarose gel to assess correct amplification. PCR products were isolated using the GeneJET PCR Purification kit (Thermo Fisher Scientific, Vilnius, Lithuania) following the manufacturer’s procedures and sent to Eurofins Genomics (Louisville, KY) for Sanger sequencing. Raw sequences were compared using BioEdit [29] and Multalin [30] software.
3.6. Data Analysis
In order to understand the relationship between the HIS1 genotypes and the presence of the Clearfield ALSS653N traits, the observed allele frequencies of both variables were subjected to a Chi-square (ꭓ2) test at significance level α = 0.05. The ꭓ2 test was performed in JMP statistical software (SAS Institute Inc., Cary, NC, USA).
4. Conclusions
In this study, we have designed a KASP assay for genotyping 37 accessions of weedy rice for HIS1 and the ALSS653N traits. In addition, we have linked the susceptibility of those weedy rice accessions to benzobicyclon (a HPPD-inhibiting herbicide). We have demonstrated a significant relationship between both the HIS1 and the ALSS653N traits. Furthermore, our results suggest that the genetic origin of weedy rice would play a role in the differential susceptibility to benzobicyclon. It is likely that weedy rice with the ALSS653N trait will surpass benzobicyclon effects.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/11/1576/s1, Table S1: Summary of the ALSS653N trait and HIS1 allele frequencies from 37 weedy rice accessions and their field response to benzobicyclon at 371 g ha−1.
Author Contributions
Conceptualization, C.B. and J.K.N.; Methodology, C.B. and F.G.-T.; Resources, J.K.N.; Writing—Original Draft Preparation, C.B. and F.G.-T.; Writing—Review & Editing, C.B., F.G.-T., and J.K.N.; Funding Acquisition, J.K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Arkansas Rice Promotion Board and by Gowan Company (Yuma, AZ, USA).
Acknowledgments
Funding for this research provided by the Arkansas Rice Research and Promotion Board and Gowan Company is greatly appreciated.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Ottis, B.V.; Smith, K.L.; Scott, R.C.; Talbert, R.E. Rice yield and quality as affected by cultivar and red rice (Oryza sativa) density. Weed Sci. 2005, 53, 499–504. [Google Scholar] [CrossRef]
- Sudianto, E.; Beng-Kah, S.; Ting-Xiang, N.; Saldain, N.E.; Scott, R.C.; Burgos, N.R. Clearfield® rice: Its development, success, and key challenges on a global perspective. Crop Prot. 2013, 49, 40–51. [Google Scholar] [CrossRef]
- Smith, R.J., Jr. Weed thresholds in Southern U.S. rice, Oryza sativa. Weed Technol. 1988, 2, 232–241. [Google Scholar] [CrossRef]
- Karn, E.; De Leon, T.; Espino, L.; Al-Khatib, K.; Brim-DeForest, W. Effects of competition from California weedy rice (Oryza sativa f. spontanea) biotypes on a cultivated rice variety. Weed Technol. 2020, 1–29, In Press. [Google Scholar] [CrossRef]
- Burgos, N.R.; Norsworthy, J.K.; Scott, R.C.; Smith, K.L. Red rice (Oryza sativa) status after 5 years of imidazolinone-resistant rice technology in Arkansas. Weed Technol. 2008, 22, 200–208. [Google Scholar] [CrossRef]
- Croughan, T.P. Clearfield rice: It’s not a GMO. La. Agric. 2003, 46, 24–26. [Google Scholar]
- Rosas, J.E.; Bonnecarrère, V.; Pérez de Vida, F. One-step, codominant detection of imidazolinone resistance mutations in weedy rice (Oryza sativa L.). Electron. J. Biotechnol. 2014, 17, 95–101. [Google Scholar] [CrossRef]
- Burgos, N.R.; Singh, V.; Tseng, T.M.; Black, H.; Young, N.D.; Huang, Z.; Hyma, K.E.; Gealy, D.R.; Caicedo, A.L. The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiol. 2014, 166, 1208–1220. [Google Scholar] [CrossRef]
- Komatsubara, K.; Sekino, K.; Yamada, Y.; Koyanagi, H.; Nakahara, S. Discovery and development of a new herbicide, benzobicyclon. J. Pestic. Sci. 2009, 34, 113–114. [Google Scholar] [CrossRef]
- Young, M.L.; Norsworthy, J.K.; Scott, R.C.; Bond, J.A.; Heiser, J. Benzobicyclon as a post-flood option for weedy rice control. Weed Technol. 2018, 32, 371–378. [Google Scholar] [CrossRef]
- Kwon, O.D.; Shin, S.H.; An, K.N.; Lee, I.; Min, H.K.; Park, H.G.; Shin, H.R.; Jung, H.L.; Kuk, Y. Response of phytotoxicity on rice varieties to HPPD- inhibiting herbicides in paddy rice fields. Korean J. Weed Sci. 2012, 32, 240–255. [Google Scholar] [CrossRef]
- Maeda, H.; Murata, K.; Sakuma, N.; Takei, S.; Yamazaki, A.; Karim, M.R.; Kawata, M.; Hirose, S.; Kawagishi-Kobayashi, M.; Taniguchi, Y.; et al. A rice gene that confers broad-spectrum resistance to β-triketone herbicides. Science 2019, 365, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.H.; Maeda, Y.; Sunohara, I.; Ando, M.; Oshima, M.; Kawata, H.; Yoshida, S.; Hirose, M.; Kawagishi, Y.; Taniguchi, K.; et al. Plant Having Increased Resistance or Susceptibility to 4-HPPD Inhibitor. U.S. Patent 10,544,425, 12 February 2015. [Google Scholar]
- Gealy, D.R.; Burgos, N.R.; Yeater, K.M.; Jackson, A.K. Outcrossing potential between U.S. blackhull red rice and Indica rice cultivars. Weed Sci. 2015, 63, 647–657. [Google Scholar] [CrossRef]
- Shivrain, V.K.; Burgos, N.R.; Agrama, H.A.; Lawton-Rauh, A.; Lu, B.; Sales, M.A.; Boyett, V.; Gealy, D.R.; Moldenhauer, K.A.K. Genetic diversity of weedy red rice (Oryza sativa) in Arkansas, USA. Weed Res. 2010, 50, 289–302. [Google Scholar] [CrossRef]
- Reagon, M.; Thurber, C.S.; Gross, B.L.; Olsen, K.M.; Jia, Y.; Caicedo, A.L. Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice. BMC Evol. Biol. 2010, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Young, M.L.; Norsworthy, J.K.; Sandoski, C.A.; Palhano, M.; Martin, S. Weedy rice control with benzobicyclon in rice: Is this possible? In Proceedings of the Weed Science Society, San Juan, Puerto Rico, USA, 8–11 February 2016.
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Hardke, J. Trends in Arkansas Rice Production. In Arkansas Rice Research Studies; Norman, R.J., Moldenhauer, K.A.K., Eds.; University of Arkansas Division of Agriculture Cooperative Extension Service: Little Rock, AR, USA, 2017; pp. 11–21. [Google Scholar]
- De Leon, T.B.; Karn, E.; Al-Khatib, K.; Espino, L.; Blank, T.; Andaya, C.B.; Andaya, V.C.; Brim-DeForest, W. Genetic variation and possible origins of weedy rice found in California. Ecol. Evol. 2019, 9, 5835–5848. [Google Scholar] [CrossRef]
- Akasaka, M.; Watanabe, H.; Kawana, Y. Inheritance for sensitivity of high-yielding rice cultivars, ‘Momiroman’ and ‘Takanari’, to benzobicyclon, a 4-HPPD inhibitor. J. Weed Sci. Technol. 2011, 56, 89–94. [Google Scholar] [CrossRef][Green Version]
- Kim, S.-Y.; Oh, S.-H.; Lee, J.-Y.; Yeo, U.-S.; Lee, J.-H.; Cho, J.-H.; Song, Y.-C.; Oh, M.-K.; Han, S.-I.; Seo, W.-D.; et al. Differential sensitivity of rice cultivars to HPPD-inhibiting herbicides and their influences on rice yield. Korean J. Weed Sci. 2012, 57, 160–165. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Han, S.-I.; Lee, J.-Y.; Cho, J.-H.; Oh, S.-H.; Park, N.-B.; Son, Y.-B.; Song, Y.-C.; Nam, M.-H. Phytotoxicity of japonica × indica-type rice varieties to HPPD-inhibiting herbicides in paddy fields. Korean J. Weed Sci. 2014, 26, 310–315. [Google Scholar] [CrossRef]
- Castner, M.C.; Norsworthy, J.K.; Brabham, C.; Sandoski, C. Impact of weedy rice growth stage on the efficacy of benzobicyclon. In Proceedings of the Southern Weed Science Society, Biloxi, MS, USA, 27–30 January 2020. [Google Scholar]
- Patterson, J.A.; Norsworthy, J.K.; Lancaster, Z.D.; Beesinger, J.W.; France, O.W. Evaluating the effect of rice growth stage at application on varietal tolerance to benzobicyclon. In Proceedings of the Southern Weed Science Society, Biloxi, MS, USA, 27–30 January 2020. [Google Scholar]
- Hardke, J.; Baker, R.; Barber, R.; Bateman, N.; Butts, T.; Hamilton, M. Rice Management Guide. Available online: https://www.uaex.edu/farm-ranch/crops-commercial-horticulture/rice/2019%20Rice%20Management%20Guide.pdf (accessed on 27 April 2020).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar] [CrossRef]
- Alexandrov, N.; Tai, S.; Wang, W.; Mansueto, L.; Palis, K.; Fuentes, R.R.; Ulat, V.J.; Chebotarov, D.; Zhang, G.; Li, Z.; et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015, 43, D1023–D1027. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).