Genome-Wide Identification of WRKY Genes in Artemisia annua: Characterization of a Putative Ortholog of AtWRKY40
Abstract
1. Introduction
2. Results
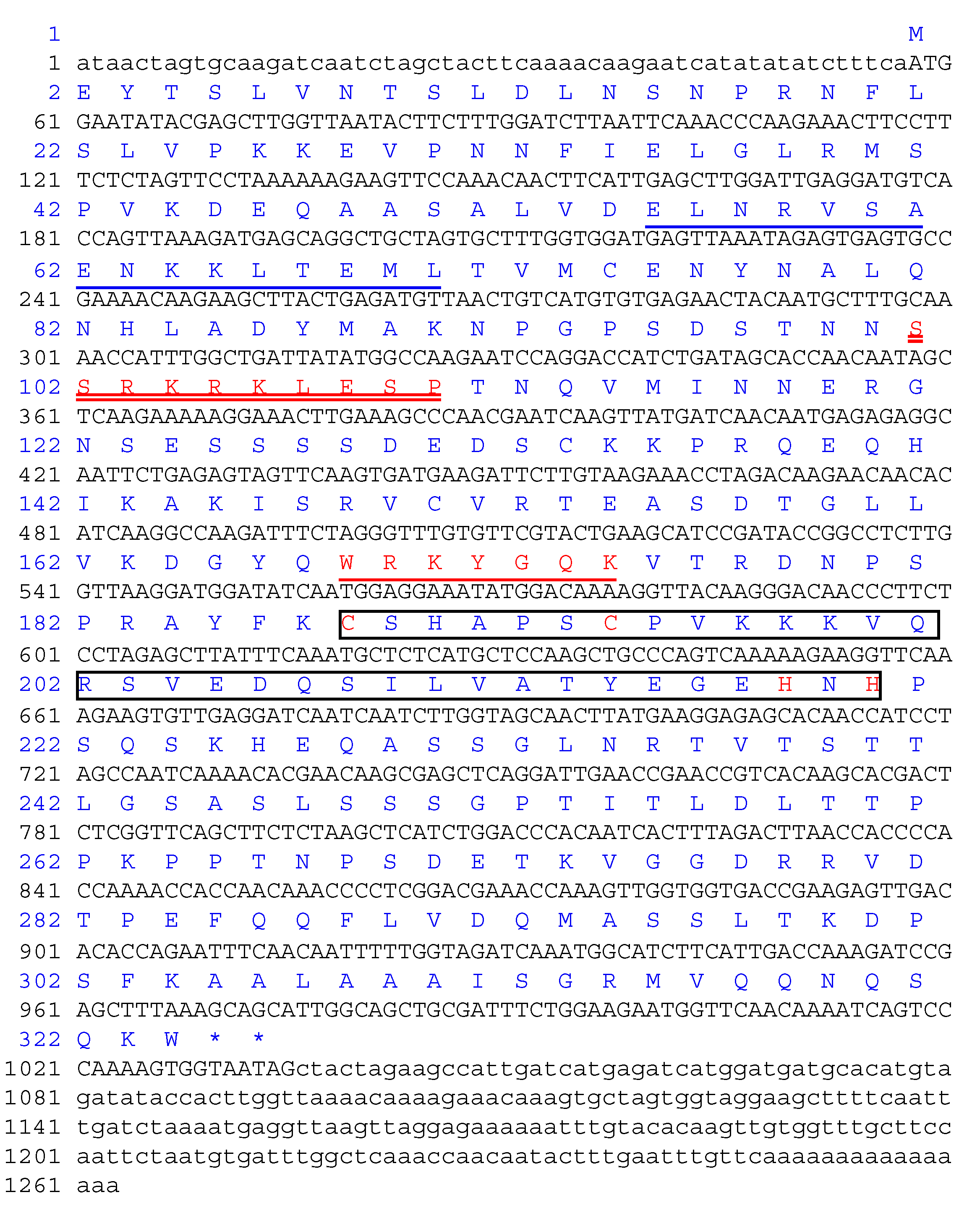
2.1. Molecular Cloning of a Novel A. annua WRKY40 Gene
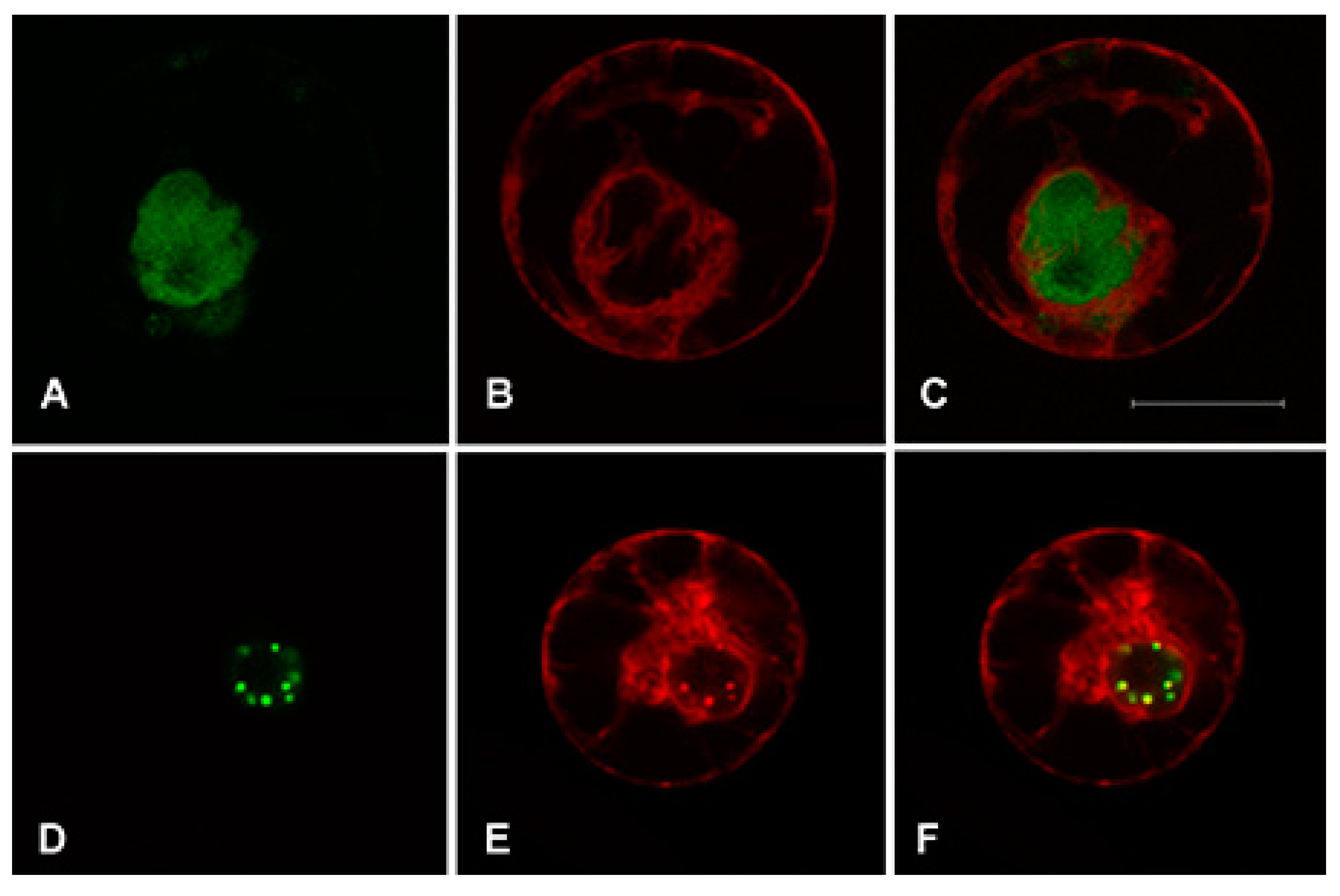
2.2. AaWRKY40 Nuclear Localization in A. annua Protoplasts
2.3. Expression Analysis in A. annua Suspension Cell Cultures
2.4. Genome-Wide Identification of WRKY Transcription Factors in A. annua
2.5. The Group IIa of WRKY Transcription Factors in A. annua
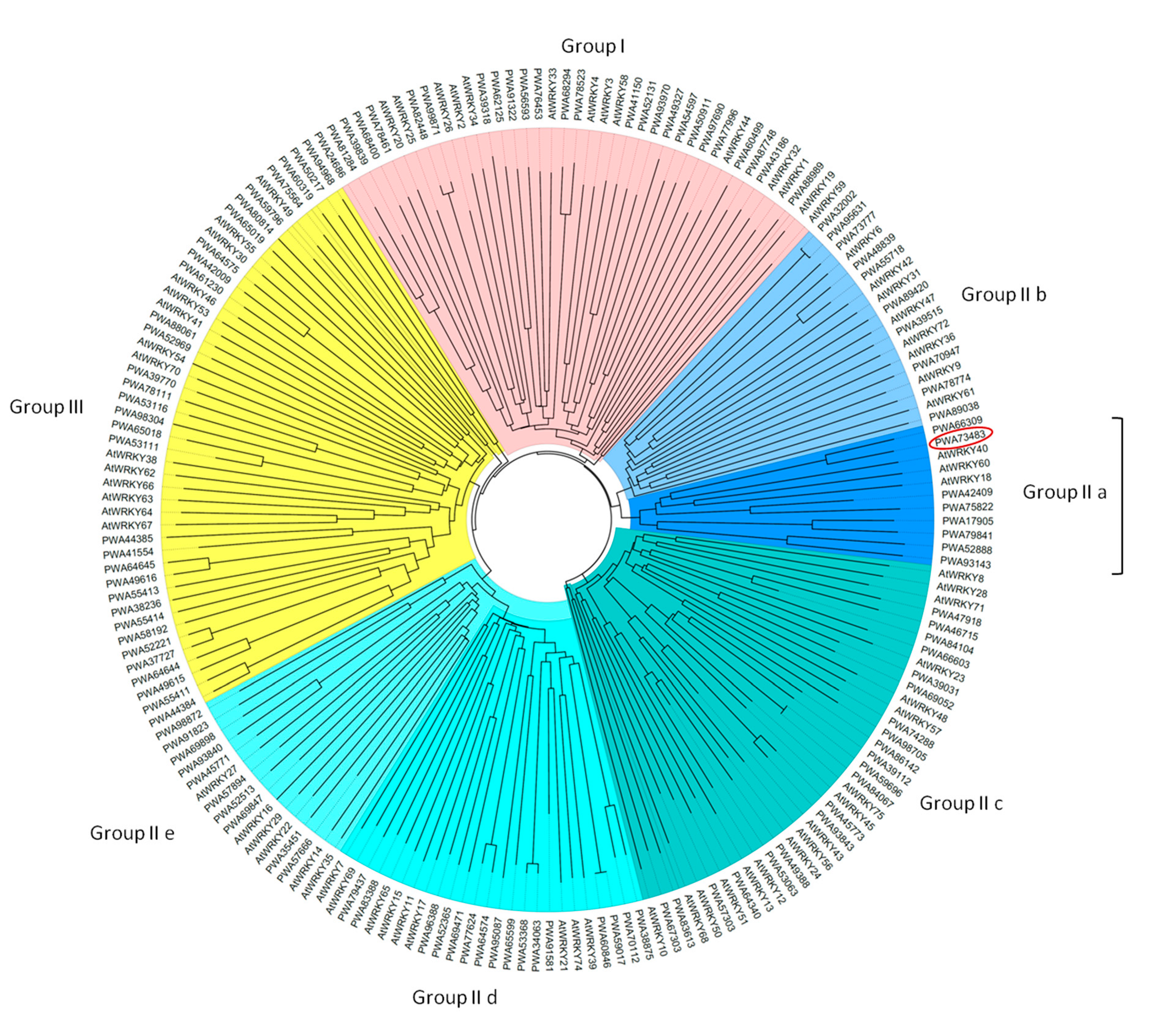
2.5.1. Phylogenetic Analysis of WRKY Group IIa of A. annua
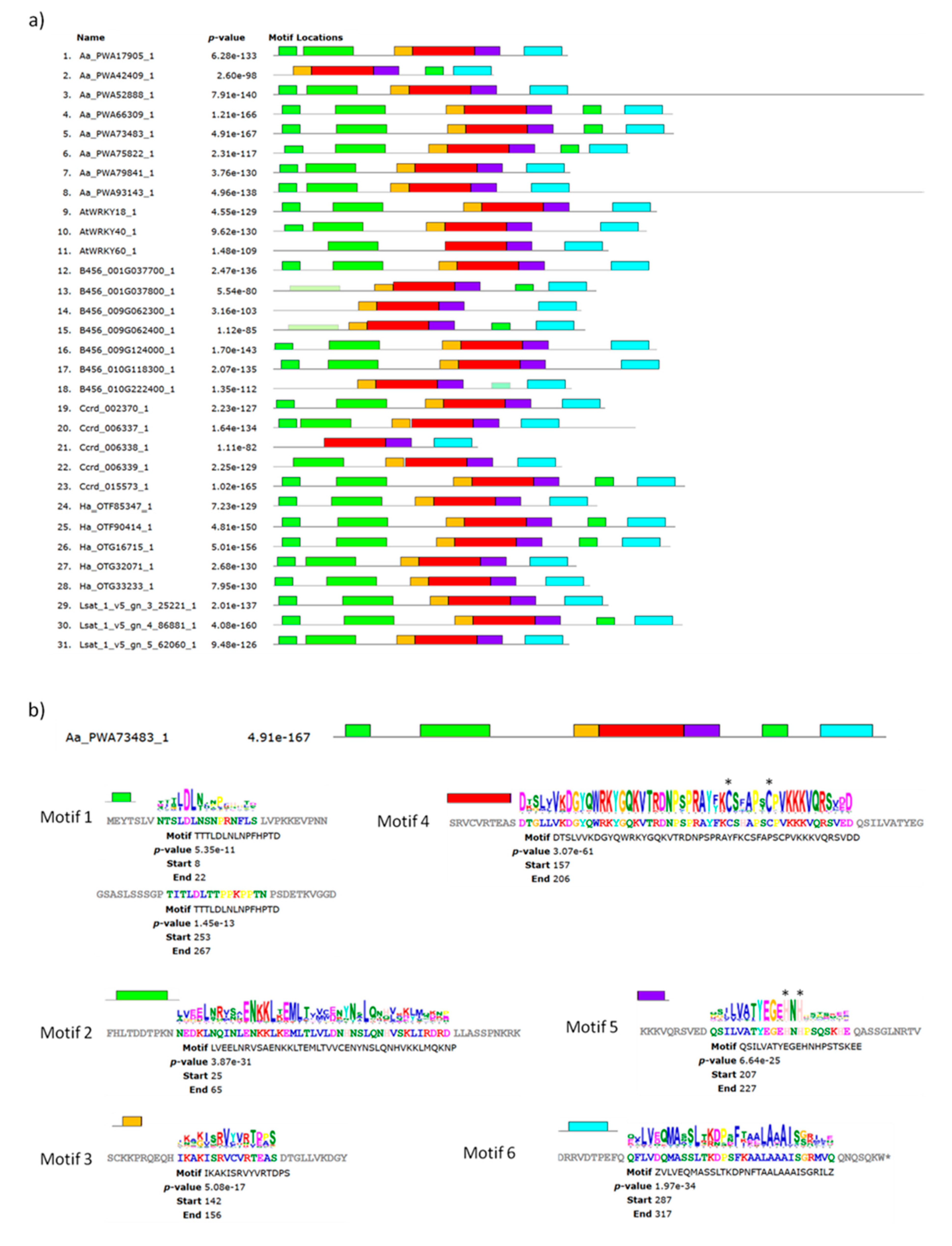
2.5.2. Conserved Protein Motifs in WRKY Group IIa of A. annua
2.6. Regulatory Motifs in the Upstream Regions of Group IIa WRKY Transcription Factors
3. Discussion
4. Materials and Methods
4.1. Plant Material and Chemical Treatments
4.2. Nucleic Acids Extraction and cDNA Preparation
4.3. Cloning and Sequence Analysis
4.4. Preparation of Gene Constructs and Protoplast Transformation
4.5. Expression Analysis of AaWRKY
4.6. Identification of WRKY Genes in the Genome of A. annua
4.7. Phylogeny, Protein Conserved Motif and Gene Structure Analysis of AaWRKY Genes
4.8. Cis-Acting Element Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant. Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- De Paolis, A.; Frugis, G.; Giannino, D.; Iannelli, M.A.; Mele, G.; Rugini, E.; Silvestri, C.; Sparvoli, F.; Testone, G.; Mauro, M.L.; et al. Plant Cellular and Molecular Biotechnology: Following Mariotti’s Steps. Plants 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. Bmc Plant. Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and b-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant. Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant. Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant. Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant. Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. Dna Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant. Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Kalde, M.; Barth, M.; Somssich, I.E.; Lippok, B. Members of the Arabidopsis WRKY Group III Transcription Factors Are Part of Different Plant Defense Signaling Pathways. Mol. Plant-Microbe Interact. 2003, 16, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Broun, P.; Liu, Y.; Queen, E.; Schwarz, Y.; Abenes, M.L.; Leibman, M. Importance of transcription factors in the regulation of plant secondary metabolism and their relevance to the control of terpenoid accumulation. Phytochem. Rev. 2006, 5, 27–38. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J.G.; Yazaki, K.; Sato, F. Identification of a WRKY Protein as a Transcriptional Regulator of Benzylisoquinoline Alkaloid Biosynthesis in Coptis japonica. Plant Cell Physiol. 2007, 48, 8–18. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Wang, J.-W.; Wang, S.; Wang, J.-Y.; Chen, X.-Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The Transcription Factor CrWRKY1 Positively Regulates the Terpenoid Indole Alkaloid Biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Guillaumie, S.; Mzid, R.; Mechin, V.; Leon, C.; Hichri, I.; Destrac-Irvine, A.; Trossat-Magnin, C.; Delrot, S.; Lauvergeat, V. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol 2010, 72, 215–234. [Google Scholar] [CrossRef]
- Weathers, P.J.; Elkholy, S.; Wobbe, K.K. Artemisinin: The biosynthetic pathway and its regulation in Artemisia annua, a terpenoid-rich species. In Vitro Cell. Dev. Biol.-Plant 2006, 42, 309–317. [Google Scholar] [CrossRef]
- Brown, G.D. The Biosynthesis of Artemisinin (Qinghaosu) and the Phytochemistry of Artemisia annua L. (Qinghao). Molecules 2010, 15, 7603–7698. [Google Scholar] [CrossRef] [PubMed]
- Caretto, S.; Quarta, A.; Durante, M.; Nisi, R.; De Paolis, A.; Blando, F.; Mita, G. Methyl jasmonate and miconazole differently affect arteminisin production and gene expression in Artemisia annua suspension cultures. Plant Biol. 2011, 13, 51–58. [Google Scholar] [CrossRef]
- Durante, M.; Caretto, S.; Quarta, A.; De Paolis, A.; Nisi, R.; Mita, G. beta-Cyclodextrins enhance artemisinin production in Artemisia annua suspension cell cultures. Appl. Microbiol. Biotechnol. 2011, 90, 1905–1913. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, F.; De Paolis, A.; Durante, M.; Blando, F.; Mita, G.; Caretto, S. Enhanced production of bioactive isoprenoid compounds from cell suspension cultures of Artemisia annua L. using beta-cyclodextrins. Int. J. Mol. Sci. 2014, 15, 19092–19105. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528. [Google Scholar] [CrossRef]
- Peplow, M. Synthetic biology’s first malaria drug meets market resistance. Nature 2016, 530, 389–390. [Google Scholar] [CrossRef]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, H.; Li, G.; et al. Isolation and Characterization of AaWRKY1, an Artemisia annua Transcription Factor that Regulates the Amorpha-4,11-diene Synthase Gene, a Key Gene of Artemisinin Biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Di Sansebastiano, G.P.; Rizzello, F.; Durante, M.; Caretto, S.; Nisi, R.; De Paolis, A.; Faraco, M.; Montefusco, A.; Piro, G.; Mita, G. Subcellular compartmentalization in protoplasts from Artemisia annua cell cultures: Engineering attempts using a modified SNARE protein. J. Biotechnol. 2015, 202, 146–152. [Google Scholar] [CrossRef]
- Geilen, K.; Bohmer, M. Dynamic subnuclear relocalization of WRKY40, a potential new mechanism of ABA-dependent transcription factor regulation. Plant Signal. Behav. 2015, 10, e1106659. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y.; et al. The Genome of Artemisia annua Provides Insight into the Evolution of Asteraceae Family and Artemisinin Biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Thilmony, R.; You, F.M.; Gu, Y.Q.; Coleman-Derr, D. PIECE 2.0: An update for the plant gene structure comparison and evolution database. Nucleic Acids Res. 2017, 45, 1015–1020. [Google Scholar] [CrossRef]
- Han, J.; Wang, H.; Lundgren, A.; Brodelius, P.E. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 2014, 102, 89–96. [Google Scholar] [CrossRef]
- Jiang, W.; Fu, X.; Pan, Q.; Tang, Y.; Shen, Q.; Lv, Z.; Yan, T.; Shi, P.; Li, L.; Zhang, L.; et al. Overexpression of AaWRKY1 Leads to an Enhanced Content of Artemisinin in Artemisia annua. Biomed. Res. Int. 2016, 2016, 9. [Google Scholar] [CrossRef]
- Chen, M.; Yan, T.; Shen, Q.; Lu, X.; Pan, Q.; Huang, Y.; Tang, Y.; Fu, X.; Liu, M.; Jiang, W.; et al. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 2017, 214, 304–316. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (Bba) Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.H.; Gadella, T.W.J., Jr.; Ferrario, S.; Busscher, M.; Angenent, G.C. Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 2002, 99, 2416–2421. [Google Scholar] [CrossRef] [PubMed]
- Faraco, M.; Di Sansebastiano, G.P.; Spelt, K.; Koes, R.E.; Quattrocchio, F.M. One Protoplast Is Not the Other! Plant Physiol. 2011, 156, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Ariani, A.; Barozzi, F.; Sebastiani, L.; di Toppi, L.S.; di Sansebastiano, G.P.; Andreucci, A. AQUA1 is a mercury sensitive poplar aquaporin regulated at transcriptional and post-translational levels by Zn stress. Plant Physiol. Biochem. 2019, 135, 588–600. [Google Scholar] [CrossRef]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 2012, 17, 349–359. [Google Scholar] [CrossRef]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; De Tommasi, N.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, Y.; Wang, Z.; Lin, L.; Cui, M.; Long, Y.; Xing, Z. Genome-Wide Identification of WRKY Transcription Factors in the Asteranae. Plants 2019, 8, 393. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. Embo J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef]
- Cormack, R.S.; Eulgem, T.; Rushton, P.J.; Köchner, P.; Hahlbrock, K.; Somssich, I.E. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim. Biophys. Acta (Bba)–Gene Struct. Expr. 2002, 1576, 92–100. [Google Scholar] [CrossRef]
- Lee, S.-J.; Kang, J.-Y.; Park, H.-J.; Kim, M.D.; Bae, M.S.; Choi, H.-I.; Kim, S.Y. DREB2C Interacts with ABF2, a bZIP Protein Regulating Abscisic Acid-Responsive Gene Expression, and Its Overexpression Affects Abscisic Acid Sensitivity. Plant Physiol. 2010, 153, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yagi, M.; Kusano, T.; Sano, H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. MGG 2000, 263, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 51, 473–497. [Google Scholar] [CrossRef]
- Nagy, J.I.; Maliga, P. Callus Induction and Plant Regeneration from Mesophyll Protoplasts of Nicotiana sylvestris. Z. Für Pflanzenphysiol. 1976, 78, 453–455. [Google Scholar] [CrossRef]
- Di Sansebastiano, G.P.; Paris, N.; Marc-Martin, S.; Neuhaus, J.M. Regeneration of a lytic central vacuole and of neutral peripheral vacuoles can be visualized by green fluorescent proteins targeted to either type of vacuoles. Plant Physiol. 2001, 126, 78–86. [Google Scholar] [CrossRef]
- Di Sansebastiano, G.-P.; Paris, N.; Marc-Martin, S.; Neuhaus, J.-M. Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 1998, 15, 449–457. [Google Scholar] [CrossRef]
- Grieco, F.; Castellano, M.A.; Di Sansebastiano, G.P.; Maggipinto, G.; Neuhaus, J.M.; Martelli, G.P. Subcellular localization and in vivo identification of the putative movement protein of olive latent virus 2. J. Gen. Virol. 1999, 80, 1103–1109. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nguyen, N.T.T.; Contreras-Moreira, B.; Castro-Mondragon, J.A.; Santana-Garcia, W.; Ossio, R.; Robles-Espinoza, C.D.; Bahin, M.; Collombet, S.; Vincens, P.; Thieffry, D.; et al. RSAT 2018: Regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res. 2018, 46, W209–W214. [Google Scholar] [CrossRef]




| seq | W-Box | G-Box | GCC-Box | TCA-Element | MeJa-RE | ERE |
|---|---|---|---|---|---|---|
| Aa_PWA66309 | 5 | 1 | 1 | 0 | 7 | 2 |
| Aa_WRKY40 | 5 | 1 | 1 | 0 | 5 | 2 |
| At_WRKY18 | 6 | 0 | 1 | 0 | 5 | 0 |
| At_WRKY40 | 5 | 0 | 0 | 0 | 5 | 1 |
| B456_001G037700 | 6 | 1 | 0 | 0 | 5 | 8 |
| Ccrd_002370 | 6 | 1 | 0 | 0 | 5 | 2 |
| Lsat_1_v5_gn_3_25221 | 7 | 0 | 0 | 0 | 5 | 0 |
| Aa_PWA93143 | 4 | 0 | 0 | 0 | 4 | 1 |
| B456_001G037800 | 7 | 1 | 1 | 0 | 4 | 1 |
| Ha_OTF90414 | 4 | 0 | 0 | 0 | 4 | 3 |
| Lsat_1_v5_gn_5_62060 | 6 | 0 | 0 | 0 | 4 | 2 |
| Aa_PWA52888 | 4 | 0 | 0 | 0 | 3 | 1 |
| Aa_PWA75822 | 3 | 0 | 0 | 0 | 3 | 2 |
| B456_009G062400 | 9 | 0 | 0 | 0 | 3 | 1 |
| Ccrd_006339 | 4 | 0 | 0 | 0 | 3 | 3 |
| Ha_OTF85347 | 4 | 0 | 0 | 0 | 3 | 7 |
| Ha_OTG16715 | 6 | 0 | 0 | 0 | 3 | 1 |
| Ha_OTG32071 | 2 | 0 | 3 | 0 | 3 | 4 |
| Ha_OTG33233 | 4 | 2 | 0 | 0 | 3 | 3 |
| Aa_PWA17905 | 6 | 0 | 0 | 0 | 2 | 0 |
| Lsat_1_v5_gn_4_86881 | 10 | 1 | 0 | 0 | 2 | 4 |
| Aa_PWA79841 | 7 | 0 | 0 | 0 | 1 | 1 |
| At_WRKY60 | 8 | 0 | 0 | 0 | 1 | 1 |
| B456_009G062300 | 6 | 1 | 0 | 0 | 1 | 4 |
| B456_010G118300 | 6 | 0 | 0 | 0 | 1 | 10 |
| B456_010G222400 | 6 | 0 | 0 | 1 | 1 | 3 |
| Ccrd_006337 | 2 | 0 | 0 | 0 | 1 | 2 |
| Ccrd_006338 | 2 | 0 | 0 | 0 | 1 | 2 |
| Aa_PWA42409 | 3 | 0 | 0 | 0 | 0 | 0 |
| B456_009G124000 | 7 | 0 | 0 | 0 | 0 | 8 |
| Ccrd_015573 | 6 | 1 | 0 | 0 | 0 | 2 |
| total | 166 | 10 | 7 | 1 | 88 | 81 |
| seq | W-Box | G-Box | GCC-Box | TCA-Element | MeJa-RE | ERE |
|---|---|---|---|---|---|---|
| AaGSW1/Aa_PWA39112 | 10 | 1 | 2 | 0 | 4 | 0 |
| AaWRKY1/Aa_PWA52969 | 8 | 1 | 0 | 0 | 4 | 2 |
| AA2132240/Aa_PWA78774 | 7 | 0 | 1 | 0 | 3 | 0 |
| Aa_PWA73483 | 5 | 1 | 1 | 0 | 5 | 2 |
| Aa_PWA66309 | 5 | 1 | 1 | 0 | 7 | 2 |
| total | 35 | 4 | 5 | 0 | 23 | 6 |
| Primer | Sequence | Application |
|---|---|---|
| AaWRKYDegR | YTTYTGICCRTAYTTICKCCA | RT-PCR |
| AaLeuDegF | GARAAYAARAARYTIACIGAR | RT-PCR |
| 3′ GSP1-F | TGGCCAAGAATCCAGGAC | 3′RACE |
| 3′ GSP2-F | GAGAGAGGCAATTCTGAG | 3′RACE |
| 5′GSP1-R | GTCCTGGATTCTTGGCCA | 5′RACE |
| 5′GSP2-R | CTCAGAATTGCCTCTCTC | 5′RACE |
| AaWRKY40-Full-F | CTAGTGCAAGATCAATCTAG | RT-PCR/Genomic |
| AaWRKY40-Full-R | CATGATCTCATGATCAATGG | RT-PCR/Genomic |
| RTWRKY40for | GGACAACCCTTCTCCTAGAGCTT | qRT-PCR |
| RTWRKY40rev | GCTCTCCTTCATAAGTTGCTACCAA | qRT-PCR |
| RTWRKY40probe | AATGCTCTCATGCTCCAAGCTGCCC | qRT-PCR |
| RTWRKY1for | GGAAACACACTTGCAACCATCA | qRT-PCR |
| RTWRKY1rev | GTGGTGGGTTGTGTTTATTTCATG | qRT-PCR |
| RTWRKY1probe | CTCGTTTGGCCGAACCACCTTTGC | qRT-PCR |
| RTUBIfor | CGGACCAGCAGAGGTTGATATT | qRT-PCR |
| RTUBIrev | CAGCCTTAAGACCAAATGGAGAGT | qRT-PCR |
| RTUBIprobe | CAGGAAAGCAGCTTGAAGATGGCCG | qRT-PCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Paolis, A.; Caretto, S.; Quarta, A.; Di Sansebastiano, G.-P.; Sbrocca, I.; Mita, G.; Frugis, G. Genome-Wide Identification of WRKY Genes in Artemisia annua: Characterization of a Putative Ortholog of AtWRKY40. Plants 2020, 9, 1669. https://doi.org/10.3390/plants9121669
De Paolis A, Caretto S, Quarta A, Di Sansebastiano G-P, Sbrocca I, Mita G, Frugis G. Genome-Wide Identification of WRKY Genes in Artemisia annua: Characterization of a Putative Ortholog of AtWRKY40. Plants. 2020; 9(12):1669. https://doi.org/10.3390/plants9121669
Chicago/Turabian StyleDe Paolis, Angelo, Sofia Caretto, Angela Quarta, Gian-Pietro Di Sansebastiano, Irene Sbrocca, Giovanni Mita, and Giovanna Frugis. 2020. "Genome-Wide Identification of WRKY Genes in Artemisia annua: Characterization of a Putative Ortholog of AtWRKY40" Plants 9, no. 12: 1669. https://doi.org/10.3390/plants9121669
APA StyleDe Paolis, A., Caretto, S., Quarta, A., Di Sansebastiano, G.-P., Sbrocca, I., Mita, G., & Frugis, G. (2020). Genome-Wide Identification of WRKY Genes in Artemisia annua: Characterization of a Putative Ortholog of AtWRKY40. Plants, 9(12), 1669. https://doi.org/10.3390/plants9121669








