Bioeffectors as Biotechnological Tools to Boost Plant Innate Immunity: Signal Transduction Pathways Involved
Abstract
1. Introduction
2. Results
2.1. Beneficial Rhizobacteria Screening: Phylogenetic Tree and Biochemical Tests
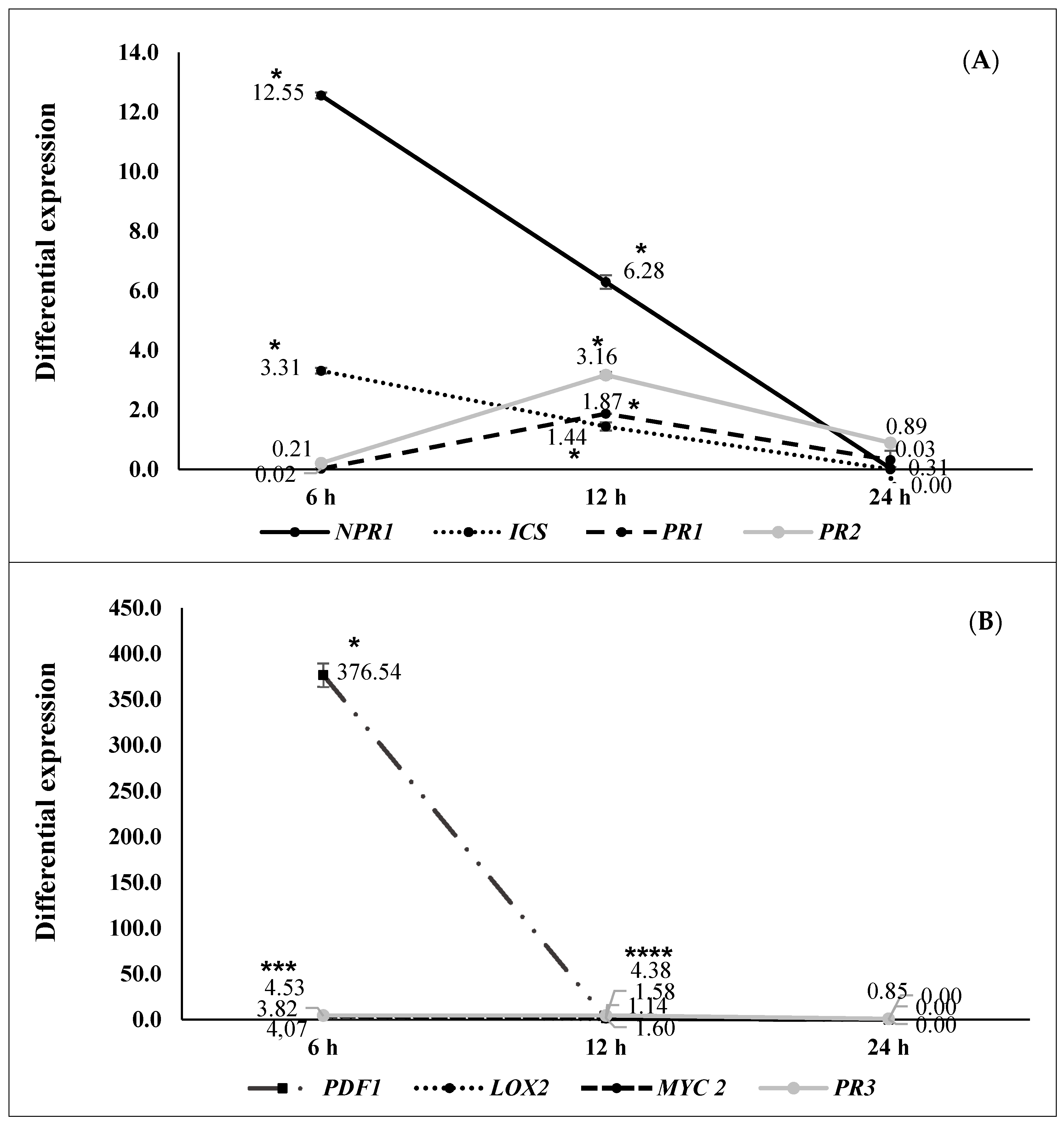
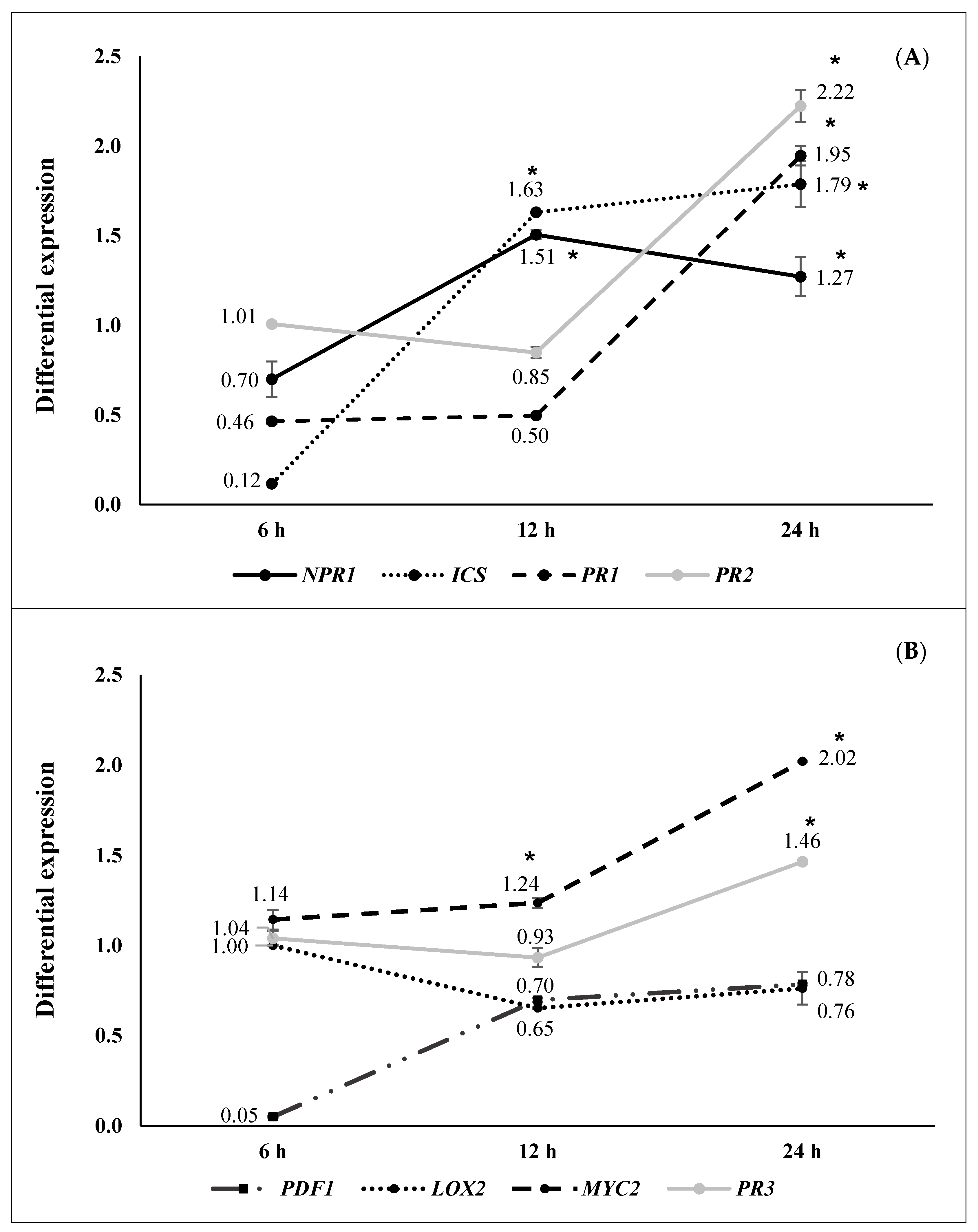
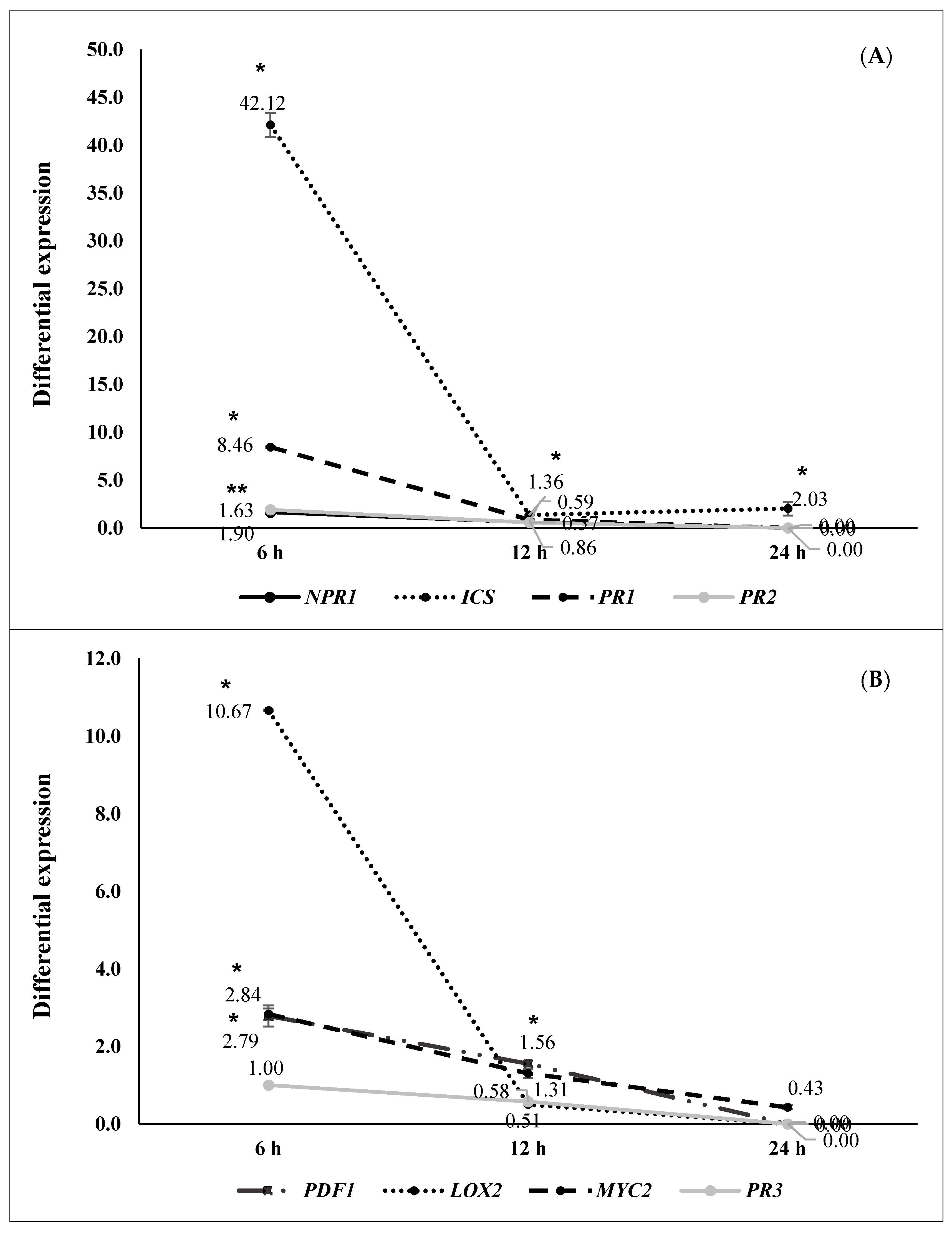
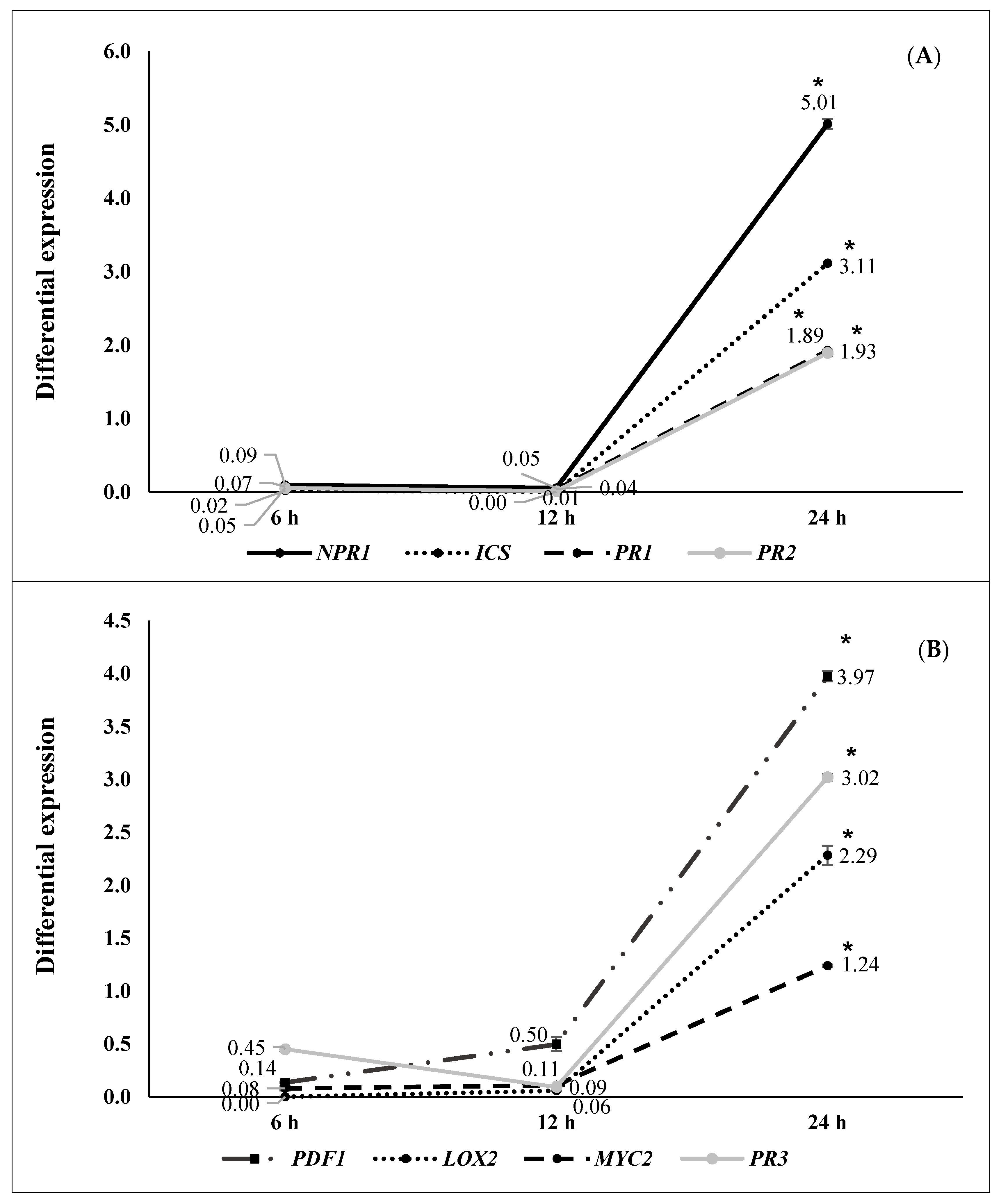
2.2. ISR by Beneficial Rhizobacteria
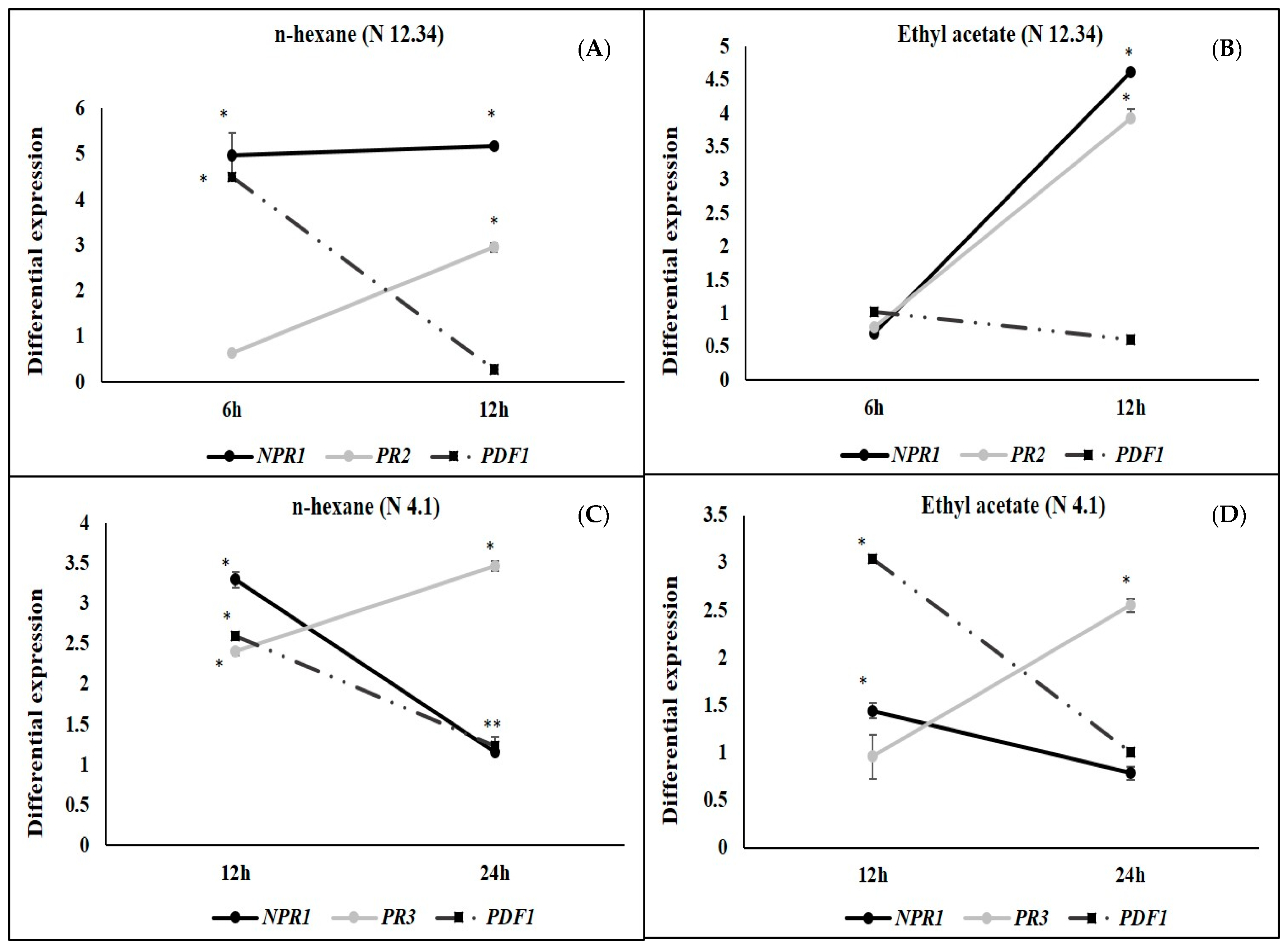
2.3. ISR by Metabolic Elicitors
3. Discussion
4. Material and Methods
4.1. Origin of Bacteria
4.2. 16S rRNA Partial Sequencing Phylogenetic Analysis
4.3. Phylogenetic Tree
4.4. Biochemical Tests for Putative Beneficial Rhizobacteria Traits
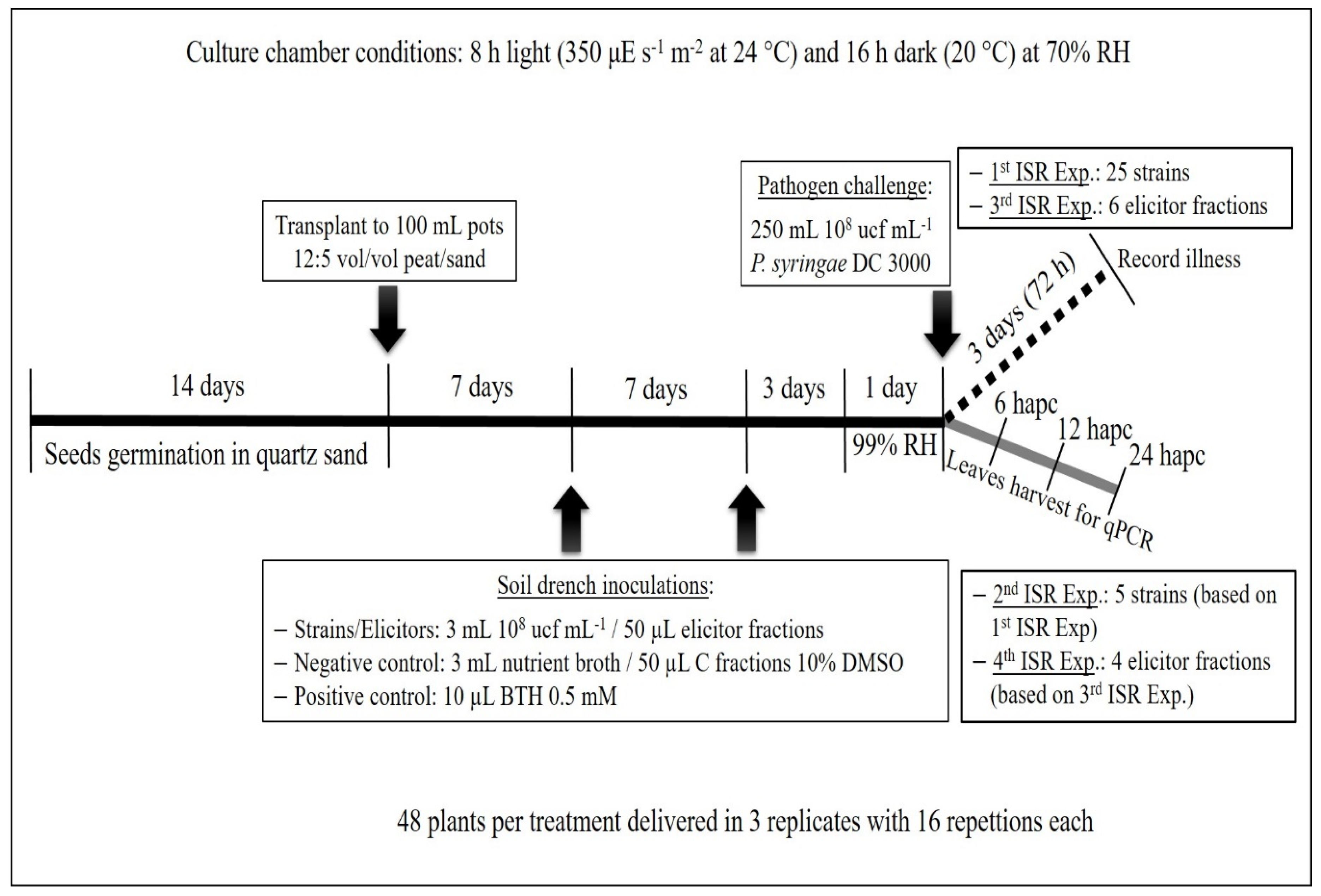
4.5. First ISR Experiment. Screening for Isolates Able to Induce Systemic Resistance
4.6. Second ISR Experiment. Study of the Signal Transduction Pathway Involved in Plant Protection
4.7. RNA Extraction and RT-qPCR Analysis (Second ISR Experiment)
4.8. Metabolic Elicitors’ Extraction and Its Capacity to Induce Systemic Resistance. Third ISR Experiment
4.9. RT-qPCR Analysis of the Genes Triggered by Metabolic Elicitor Fractions (Fourth ISR Experiment)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Availability of Materials and Data
Abbreviations
| BTH | Benzothiadiazole |
| DMSO | Dimethyl sulfoxide |
| ET | Ethylene |
| ETI | Effector-triggered immunity |
| hapc | Hours after pathogen challenge |
| IAA | Indole acetic acid |
| ICS | Isochorismate synthase |
| ISR | Induced systemic resistance |
| JA | Jasmonic acid |
| LOX 2 | Lipoxygenase 2 |
| MAMP | Microbe-associated molecular pattern |
| NPR1 | Nonexpressor of pathogenesis-related protein 1 |
| PAMP | Pathogen-associated molecular pattern |
| PDF1 | Plant defensin 1 |
| PR1 | Pathogenesis-related gene 1 |
| PRR | Pattern recognition receptor |
| PTI | Pattern-triggered immunity |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
References
- Pechanova, O.; Pechan, T. Maize-pathogen interactions: An ongoing combat from a proteomics perspective. Int. J. Mol. Sci. 2015, 16, 28429–28448. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.N.G.; Alves, G.S.C.; Van Sluys, M.-A. Plant immunity: Unravelling the complexity of plant responses to biotic stresses. Ann. Bot. 2017, 119, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Pel, M.J.; Pieterse, C.M. Microbial recognition and evasion of host immunity. J. Exp. Bot. 2013, 64, 1237–1248. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Pel, M.J.C. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Hammerschmidt, R. Systemic acquired resistance. Adv. Bot. Res. 2009, 51, 173–222. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Pelt, J.A.; Tona, J.; Parchmannb, S.; Mueller, M.J.; Buchala, A.J.; Métrauxc, J.-P.; Van Loon, L.C. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant. Pathol. 2000, 57, 123–134. [Google Scholar] [CrossRef]
- Gozzo, F.; Faoro, F. Systemic Acquired Resistance (50 Years after Discovery): Moving from the Lab to the Field. J. Agric. Food Chem. 2013, 61, 12473–12491. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Loon, L.C. NPR1: The spider in the web of induced resistance signalling pathways. Curr. Opin. Plant. Biol. 2004, 7, 456–464. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Loon, L.C. Signalling Cascades Involved in Induced Resistance. In Induced Resistance for Plant Defence; Walters, D., Newton, A.C., Lyon, G., Eds.; Blackwell: London, UK, 2007; pp. 65–88. [Google Scholar]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of Ethylene-Response-Factor1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. Ethylene response factor integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M.J. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Pieterse, C.M.J.; Mauch-Mani, B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shetn, Q.-R.; Zhalng, R. Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef]
- Garcia, J.A.L.; Probanza, A.; Ramos, B.; Mañero, F.J.G. Genetic variability of rhizobacteria from wild populations of four Lupinus species based on PCR-RAPDs. J. Plant Nutr. Soil Sci. 2001, 164, 1–7. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Kirstin Feussner, K.Y.; Sietske Van Bentum, R.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Ramos-Solano, B.; Lucas, J.A.; Garcia-Villaraco, A.; Algar, E.; Garcia-Cristobal, J.; Gutiérrez-Mañero, F.J. Siderophore and chitinase producing isolates from the rhizosphere of Nicotiana glauca Graham enhance growth and induce systemic resistance in Solanum lycopersicum L. Plant Soil 2010, 334, 189–197. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defence. Ann. Rev. Plant Biol. 2014, 66, 487–511. [Google Scholar] [CrossRef]
- Sergeeva, E.; Danielle Hirkala, L.M.; Louise, N.M. Production of indole-3-acetic acid, aromatic amino acid aminotransferase activities and plant growth promotion by Pantoea agglomerans rhizosphere isolates. Plant Soil 2007, 297, 1–13. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizo- sphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- De Freitas, J.; Banerjee, M.; Germida, J. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.). Biol. Fertil. Soils 1997, 24, 358–364. [Google Scholar] [CrossRef]
- Rodríguez-Kábana, R.; Godoy, G.; Morgan-Jones, G.; Shelby, R.A. The determination of soil chitinase activity: Conditions for assay and ecological studies. Plant Soil 1983, 75, 95–106. [Google Scholar] [CrossRef]
- Frändberg, E.; Shnurer, J. Antifungal activity of chitino- lytic bacteria isolated from airtight stored cereal grain. Can. J. Microbiol. 1998, 44, 121–127. [Google Scholar] [CrossRef]
- Sumayo, M.; Hahm, M.-S.; Ghim, S.-Y. Determinants of Plant Growth-promoting Ochrobactrum lupini KUDC1013 Involved in Induction of Systemic Resistance against Pectobacterium carotovorum subsp. carotovorum in Tobacco Leaves. Plant Pathol. J. 2013, 29, 174–181. [Google Scholar] [CrossRef]
- Lucas, J.A.; Garcia-Villaraco, A.; Ramos, B.; Garcia-Cristobal, J.; Algar, E.; Gutierrez-Mañero, J. Structural and functional study in the rhizosphere of Oryza sativa L. plants growing under biotic and abiotic stress. J. Appl. Microbiol. 2013, 115, 218–235. [Google Scholar] [CrossRef]
- Aarab, S.; Ollero, F.J.; Megías, M.; Laglaoiu, A.; Bakkali, M.; Arakrak, A. Isolation and screening of bacteria from rhizospheric soils of rice fields in Northwestern Morocco for different plant growth promotion (PGP) activities: An in vitro study. Int. J. Curr. Microbiol. App. Sci. 2015, 4, 260–269. [Google Scholar]
- Anwar, S.; Ali, B.; Sajid, I. Screening of Rhizospheric Actinomycetes for Various In-vitro and In-vivo Plant Growth Promoting (PGP) Traits and for Agroactive Compounds. Front. Microbiol. 2016, 7, 82. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Guo, X.; Zhang, Y.; Ye, B. Comparative analysis of bacterial community structure in the rhizosphere of maize by high throughput pyrosequencing. PLoS ONE 2017, 12, e178425. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Gao, J.; Wang, X.; Fan, F.; Ma, X.; Yin, H.; Zhang, C.; Feng, K.; Deng, Y. Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol. Biochem. 2017, 104, 208–217. [Google Scholar] [CrossRef]
- Raymond, K.; Muller, G.; Matzanke, B. Complexation of iron by siderophores a review of their solution and structural chemistry and biological function. Top. Curr Chem. 1984, 123, 49–102. [Google Scholar] [CrossRef]
- Jin, C.W.; He, Y.F.; Tang, C.X.; Wu, P.; Zheng, S.J. Mechanisms of microbially enhanced Fe acquisition in red clover (Trifolium pratense L.). Plant. Cell Environ. 2006, 29, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, S.J.; Johnson, R.; Hamill, J.D. Analysis of would- induced gene expression in Nicotiana species with contrasting alkaloid profiles. Functional Plant Biol. 2004, 31, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Barriuso, J.; Ramos Solano, B.; Fray, R.G.; Cámara, M.; Hartmann, A.; Gutiérrez Mañero, F.J. Transgenic tomato plants alter quorum sensing in Plant Growth Promoting Rhizo- bacteria. Plant Biotechnol J. 2008, 6, 442–452. [Google Scholar] [CrossRef]
- Sid, A.; Ezziyyani, M.; Egea-Gilabert, C.; Candela, M.E. Selecting bacterial strains for use in the biocontrol of diseases caused by Phytophthora capsici and Alternaria alteranata in sweet pepper plants. Biol. Plant 2003, 47, 569–574. [Google Scholar] [CrossRef]
- Adesina, M.F.; Lembke, A.; Costa, R.; Speksnijder, A.; Smalla, K. Screening of bacterial isolates from various European soils for in vitro antagonistic activity towards Rhizoctonia solani and Fusarium oxysporum: Site-dependent composition and diversity revealed. Soil Biol. Biochem. 2007, 39, 2818–2828. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Bakker, P.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef]
- Ramamoorthy, V.; Viswanathan, R.; Raguchander, T.; Prakasam, V.; Samiyappan, R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop. Prot. 2001, 20, 1–11. [Google Scholar] [CrossRef]
- Solano, B.R.; Barriuso, J.; Pereyra, M.T.; Domenech, J.; Mañero, F.J.G. Systemic disease protection elicited by plant growth promoting rhizobacteria strains: Relationship between metabolic responses, systemic disease protection and biotic elicitors. Phytopathoogy 2008, 98, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.; Reiber, K.; Ali, S.S.; Berney, M.; Doohan, F.M. Auxin as a player in the biocontrol of Fusarium head blight disease of barley and its potential as a disease control agent. BMC Plant Biol. 2012, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Akram, W.; Anjum, T.; Ali, B. Phenylacetic acid is ISR determinant produced by Bacillus fortis IAGS162, which involves extensive re-modulation in metabolomics of tomato to protect against Fusarium Wilt. Front. Plant Sci. 2016, 7, 498. [Google Scholar] [CrossRef]
- Martin-Rivilla, H.; Gutierrez-Mañero, F.J.; Gradillas, A.P.; Navarro, M.O.; Andrade, G.; Lucas, J.A. Identifying the compounds of the metabolic elicitors of Pseudomonas fluorescens N 21.4 responsible for their ability to induce plant resistance. Plants 2020, 9, 1020. [Google Scholar] [CrossRef]
- Kannojia, P.; Choudhary, K.K.; Srivastava, A.K.; Singh, A.K. Chapter Four. PGPR Bioelicitors: Induced Systemic Resistance (ISR) and proteomic perspective on biocontrol. In PGPR Amelioration in Sustainable Agriculture; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 67–84. [Google Scholar]
- Gutierrez Albanchez, E.; Garcia-Villaraco, A.; Lucas, J.A.; Gutierrez, F.J.; Ramos-Solano, B. Priming fingerprint induced by Bacillus amyloliquefaciensQV15, a common pattern in Arabidopsis thaliana and in field-grown blackberry. J. Plant Interact. 2018, 13, 398–408. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Spoel, S.H.; De Lange, E.S.; Abe, H.; Kobayashi, M.; Tsuda, S.; Millenaar, F.F.; Welschen, R.A.M.; Ritsema, T.; Pieterse, C.M.J. Ethylene modulates the role of nonexpressor of pathogenesis-related genes1 in cross talk between salicylate and jasmonate signalling. Plant Physiol. 2009, 149, 1797–1809. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Liu, L.; Huang, N.; Wang, L.; Ling, H.; Sun, T.; Ahmad, W.; Muhammad, K.; Guo, J.; Xu, L.; Gao, S.; et al. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic acid and Jasmonic acid pathways are activated in spatially different domains around the infection site during Effector-Triggered Immunity in Arabidopsis thaliana. Plant Cell Physiol. 2017, 59, 8–16. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant Immunity: From signalling to epigenetic control of defense. Trends Plant Sci. 2018, 9, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Munhoz, L.D.; Fonteque, J.P.; Santos, I.M.O.; Navarro, M.O.P.; Simionato, A.S.; Goya Rezende, M.I.; Balbi-Peña, M.I.; de Oliveira, A.G.; Andrade, G. Control of bacterial stem rot on tomato by extracellular bioactive compounds produced by Pseudomonas aeruginosa LV strain. Cogent Food Agric. 2017, 35, 1282592. [Google Scholar] [CrossRef]
- Martin-Rivilla, H.; Garcia-Villaraco, A.; Ramos-Solano, B.; Gutierrez-Manero, F.J.; Lucas, J.A. Extracts from cultures of Pseudomonas fluorescens induce defensive patterns of gene expression and enzyme activity while depressing visible injury and reactive oxygen species in Arabidopsis thaliana challenged with pathogenic Pseudomonas syringae. AoB Plants 2019, 11, plz049. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rivilla, H.; Garcia-Villaraco, A.; Ramos-Solano, B.; Gutierrez-Manero, F.J.; Lucas, J.A. Improving flavonoid metabolismin blackberry leaves and plant fitness by using the bioeffector Pseudomonas fluorescens N 21.4 and its metabolic elicitors: A biotechnological approach for a more sustainable crop. J. Agric. Food Chem. 2020, 68, 6170–6180. [Google Scholar] [CrossRef]
- Martin-Rivilla, H.; Garcia-Villaraco, A.; Ramos-Solano, B.; Gutierrez-Manero, F.J.; Lucas, J.A. Metabolic elicitors of Pseudomonas fluorescens N 21.4 elicit flavonoid metabolism in blackberry fruit. J. Sci. Food Agric. 2020, 101, 205–214. [Google Scholar] [CrossRef]
- Fatima, S.; Anjum, T. Identification of a potential ISR determinant from Pseudomonas aeruginosa PM12 against Fusarium Wilt in Tomato. Front. Plant Sci. 2017, 8, 69. [Google Scholar] [CrossRef]
- Van Hulten, M.; Pelser, M.; van Loon, L.C.; Pieterse, C.M.J.; Ton, J. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 5602–5607. [Google Scholar] [CrossRef]
- Lucas, J.A.; Garcia-Cristobal, J.; Bonilla, A.; Ramos, B.; Gutiérrez-Mañero, J. Beneficial rhizobacteria from rice rhizosphere confers high protection against biotic and abiotic stress inducing systemic resistance in rice seedlings. Plant Physiol. Biochem. 2014, 82, 44–53. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.-C. Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef]
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. corrigendum: Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2002, 417, 571. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Liu, H.X.; Jiang, C.H.; Wang, Y.P.; Wang, Q.Y.; Jin, H.L.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate and jasmonate/ethylene-dependent signaling pathways. Mol. Plant Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef]
- Seyfferth, C.; Tsuda, K. Salicylic acid signal transduction: The initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 2014, 5, 697. [Google Scholar] [CrossRef]
- Nie, P.; Li, X.; Wang, S.; Guo, J.; Zhao, H.; Niu, D. Induced Systemic Resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-Dependent Signalling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 238. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- Kazan, K. A new twist in SA signalling. Nature Plants 2018, 4, 327–328. [Google Scholar] [CrossRef]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Pangesti, N.; Pineda, A.; Dicke, M.; Van Loon, J.J.A. Variation in plant-mediated interactions between rhizobacteria and caterpillars: Potential role of soil composition. Plant Biol. 2014, 17, 474–483. [Google Scholar] [CrossRef]
- Du, M.; Zhao, J.; Tzeng, D.T.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Jeandet, P.; Clément, C.; Courot, E.; Cordelier, S. Modulation of Phytoalexin Biosynthesis in Engineered Plants for Disease Resistance. Int. J. Mol. Sci. 2013, 14, 14136–14170. [Google Scholar] [CrossRef] [PubMed]
- Lemarié, S. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168. [Google Scholar] [PubMed]
- Jiang, C.-H.; Fan, Z.-H.; Xie, P.; Guo, J.-H. Bacillus cereus AR156 extracellular polysaccharides served as a novel micro-associated molecular pattern to induced systemic immunity to Pst DC3000 in Arabidopsis. Front. Microbiol. 2016, 7, 977. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.; Arraes, F.B.M.; Campos, M.A.; Grossi-de-Sa, M.; Fernandez, D.; Candido, E.S.; Cardoso, M.H.; Franco, O.L.; Grossi-de-Sa, M.F. Review: Potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci. 2018, 270, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. Introducción a la Bioestadística; Editorial Reverte, S.A., Ed.; Editorial Reverte: Barcelona, Spain, 1980; p. 362. [Google Scholar]


| Gram-Negative Group | ||||||||
| Biochemical Traits | Serratia | Enterobacter | Pantoea | Erwinia | Cronobacter | Acinetobacter | Pseudomonas | Stenotrophomonas |
| Indole acetic acid (IAA) production | 0.00 | 37.5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Siderophores production | 100.00 | 87.5 | 100.00 | 100.00 | 100.00 | 83.33 | 100.00 | 100.00 |
| Phosphate solubilization | 0.00 | 75.00 | 45.45 | 50.00 | 0.00 | 100.00 | 91.67 | 0.00 |
| Chitinases production | 0.00 | 0.00 | 0.00 | 0.00 | 0,00 | 0.00 | 2.08 | 100.00 |
| Siderophores production and phosphate solubilization | 0.00 | 62.50 | 45.45 | 50.00 | 0.00 | 83.33 | 85.42 | 0.00 |
| Siderophores and chitinases production | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.08 | 100.00 |
| Siderophores and IAA production and phosphate solubilization | 0.00 | 25.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Gram-Positive Group | ||||||||
| Biochemical Traits | Bacillus cereus | Bacillus pumillus | Bacillus subtilis | Brevibacterium sp. | Bacillus endophyticus | Bacillus megaterium | ||
| IAA production | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Siderophores production | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | ||
| Phosphate solubilization | 23.08 | 0.00 | 0.00 | 11.76 | 0.00 | 36.84 | ||
| Chitinases production | 46.15 | 0.00 | 20.00 | 0.00 | 0.00 | 0.00 | ||
| Siderophores production and phosphate solubilization | 23.08 | 0.00 | 0.00 | 11.76 | 0.00 | 36.84 | ||
| Siderophores and chitinases production | 15.38 | 0.00 | 20.00 | 0.00 | 0.00 | 0.00 | ||
| Siderophores and chitinases production and phosphate solubilization | 7.69 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Biochemical Traits | ||||||
|---|---|---|---|---|---|---|
| Bacterial Strain | IAA Production | Siderophores Production | Chitinases Production | Phosphate Solubilization | ||
| GRAM− | N 5.12 | Pseudomonas putida | + | + | ||
| N 8.17 | Stenotrophomonas maltophilia | + | + | |||
| N 8.22 | Stenotrophomonas sp. | + | + | |||
| N 9.11 | Pseudomonas reinekei | + | + | |||
| N 10.6 | Pseudomonas putida | + | + | |||
| N 10.7 | Serratia odorifera | + | ||||
| N 10.21 | Pseudomonas putida | + | + | |||
| N 12.34 | Serratia rubidaea | + | ||||
| N 15.23 | Pseudomonas brassicacearum | + | + | |||
| N 16.3 | Pantoea sp. | + | + | |||
| N 16.15 | Enterobacter sp. | + | + | |||
| N 16.23 | Pantoea agglomerans | + | + | |||
| N 16.24 | Enterobacter sp. | + | + | + | ||
| N 18.10 | Pseudomonas fragi | + | + | |||
| N 21.24 | Pseudomonas fluorescens | + | + | |||
| GRAM+ | N 4.1 | Bacillus cereus | + | + | ||
| N 5.20 | Bacillus cereus | + | + | + | ||
| N 8.10 | Bacillus sp. | + | + | |||
| N 11.5 | Brevibacterium sp. | + | + | |||
| N 11.14 | Bacillus endophyticus | + | ||||
| N 11.20 | Bacillus atrophaeus | + | + | |||
| N 11.22 | Bacillus megaterium | + | + | |||
| N 11.36 | Bacillus megaterium | + | + | |||
| N 11.40 | Bacillus megaterium | + | + | |||
| N 20.15 | Bacillus simplex | + | + | |||
| Treatment | % of Protection | ||
|---|---|---|---|
| Controls | Negative Control | Nutrient Broth | 0 |
| Positive Control | Benzothiadiazole (BTH) | 54.21 ± 4.03 * | |
| GRAM− Strains | N 5.12 | Pseudomonas putida | 57.69 ± 1.76 * |
| N 8.17 | Stenotrophomonas maltophilia | 64.87 ± 1.79 * | |
| N 8.22 | Stenotrophomonas sp. | 0 | |
| N 9.11 N 10.6 | Pseudomonas reinekei Pseudomonas putida | 51.44 ± 6.88 * | |
| 0 | |||
| N 10.7 N 10.21 | Serratia odorífera Pseudomonas putida | 33.76 ± 3.22 * 0 | |
| N 12.34 | Serratia rubidaea | 56.64 ± 2.15 * | |
| N 15.23 | Pseudomonas brassicacearum | 0 | |
| N 16.3 | Pantoea sp. | 14.91 ± 2.45 * | |
| N 16.15 | Enterobacter sp. | 24.18 ± 1.96 * | |
| N 16.23 | Pantoea agglomerans | 21.21 ± 7.32 * | |
| N 16.24 | Enterobacter sp. | 6.93 ± 2.31 | |
| N 18.10 | Pseudomonas fragi | 0 | |
| N 21.24 | Pseudomonas fluorescens | 82.08 ± 2.46 * | |
| GRAM+ Strains | N 4.1 | Bacillus cereus | 69.45 ± 0.38 * |
| N 5.20 | Bacillus cereus | 49.75 ± 0.82 * | |
| N 8.10 | Bacillus sp. | 22.93 ± 2.93 * | |
| N 11.5 | Brevibacterium sp. | 29.82 ± 1.82 * | |
| N 11.14 | Bacillus endophyticus | 0 | |
| N 11.20 | Bacillus atrophaeus | 42.72 ± 3.51 * | |
| N 11.22 | Bacillus megaterium | 0 | |
| N 11.36 | Bacillus megaterium | 0 | |
| N 11.40 | Bacillus megaterium | 23.98 ± 0.18 * | |
| N 20.15 | Bacillus simplex | 30.83 ± 4.92 * | |
| Treatment | % of Protection | |
|---|---|---|
| Controls | Negative control | 0 |
| Positive control (BTH) | 52.09 ± 1.75 * | |
| N 12.34 | n-Hexane | 61.26 ± 2.23 * |
| Ethyl acetate | 54.64 ± 1.48 * | |
| n-Butanol | 35.42 ± 2.77 * | |
| N 4.1 | n-Hexane | 68.11 ± 0.76 * |
| Ethyl acetate | 67.30 ± 3.76 * | |
| n-Butanol | 52.31 ± 1.91 * | |
| Forward Primer | Reverse Primer | |
|---|---|---|
| AtNPR1 | 5′-TATTGTCAARTCTRATGTAGAT | 5′-TATTGTCAARTCTRATGTAGAT |
| AtPR1 | 5′-AGTTGTTTGGAGAAAGTCAG | 5′-GTTCACATAATTCCCACGA |
| AtICS | 5′-GCAAGAATCATGTTCCTACC | 5′AATTATCCTGCTGTTACGAG |
| AtPdf1 | 5′-TTGTTCTCTTTGCTGCTTTCGA | 5′-TTGGCTTCTCGCACAACTTCT |
| AtLOX2 | 5′-ACTTGCTCGTCCGGTAATTGG | 5′-GTACGGCCTTGCCTGTGAATG |
| AtMYC2 | 5′GATGAGGAGGTGACGGATACGGAA | 5′-CGCTTTACCAGCTAATCCCGCA |
| AtPR2 | 5′-TCGTCTCGATTATGCTCTCTTC | 5′-GCAGAATACACAGCATCCAAAA |
| AtPR3 | 5′-AAATCAACCTAGCAGGCCACT | 5′-GAGGGAGAGGAACACCTTGACT |
| Sand | 5′ -CTGTCTTCTCATCTCTTGTC | 5′-TCTTGCAATATGGTTCCTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin-Rivilla, H.; Garcia-Villaraco, A.; Ramos-Solano, B.; Gutierrez-Mañero, F.J.; Lucas, J.A. Bioeffectors as Biotechnological Tools to Boost Plant Innate Immunity: Signal Transduction Pathways Involved. Plants 2020, 9, 1731. https://doi.org/10.3390/plants9121731
Martin-Rivilla H, Garcia-Villaraco A, Ramos-Solano B, Gutierrez-Mañero FJ, Lucas JA. Bioeffectors as Biotechnological Tools to Boost Plant Innate Immunity: Signal Transduction Pathways Involved. Plants. 2020; 9(12):1731. https://doi.org/10.3390/plants9121731
Chicago/Turabian StyleMartin-Rivilla, Helena, Ana Garcia-Villaraco, Beatriz Ramos-Solano, Francisco Javier Gutierrez-Mañero, and Jose Antonio Lucas. 2020. "Bioeffectors as Biotechnological Tools to Boost Plant Innate Immunity: Signal Transduction Pathways Involved" Plants 9, no. 12: 1731. https://doi.org/10.3390/plants9121731
APA StyleMartin-Rivilla, H., Garcia-Villaraco, A., Ramos-Solano, B., Gutierrez-Mañero, F. J., & Lucas, J. A. (2020). Bioeffectors as Biotechnological Tools to Boost Plant Innate Immunity: Signal Transduction Pathways Involved. Plants, 9(12), 1731. https://doi.org/10.3390/plants9121731






