Identification of Cis-Regulatory Sequences Controlling Pollen-Specific Expression of Hydroxyproline-Rich Glycoprotein Genes in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Integrative Motif Discovery Pipeline for Pollen-Specific HRGPs
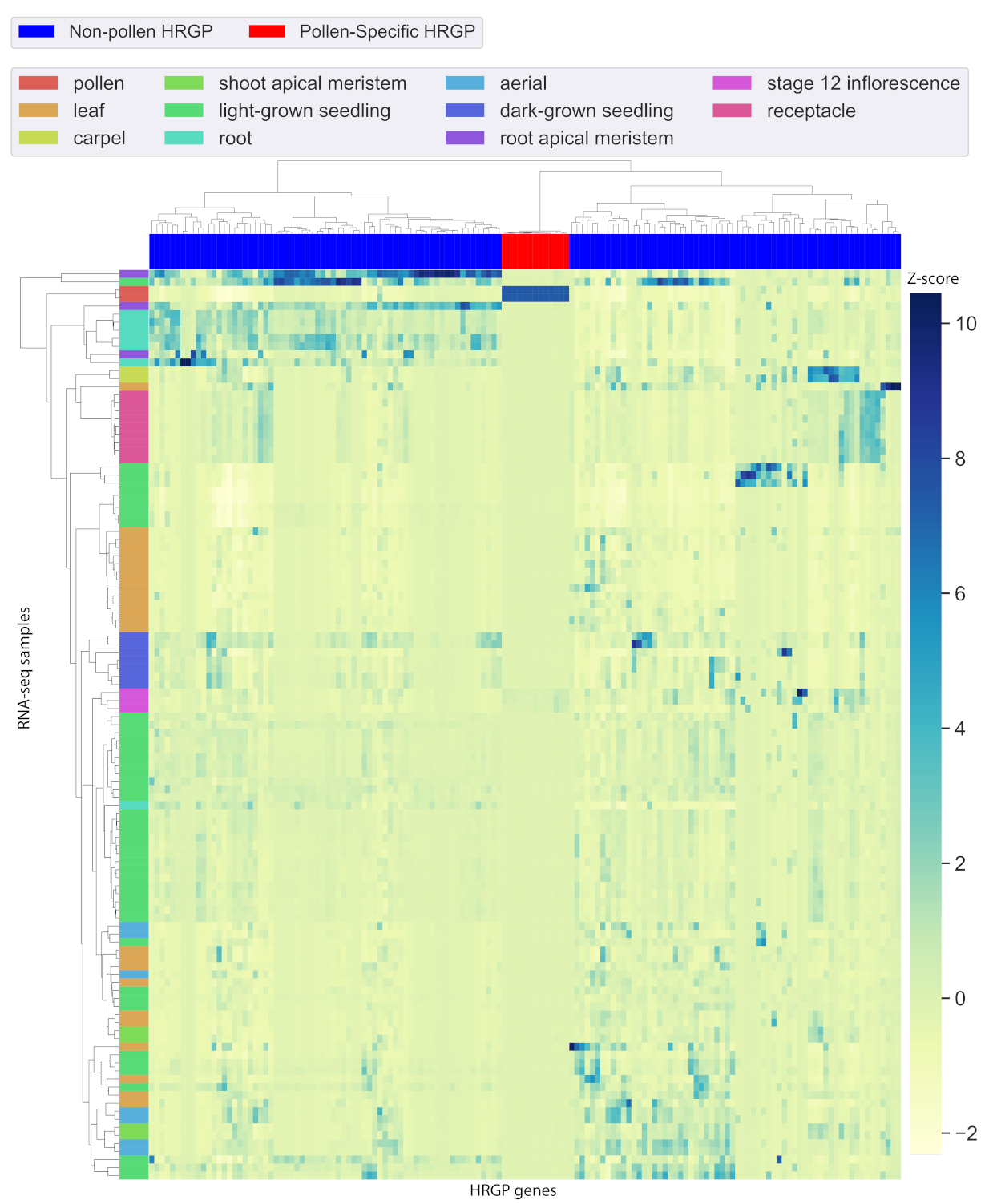
2.2. Identification of Pollen-Specific HRGP Genes
2.3. Integrative Analysis Filter A: A Relaxed Set of Pollen-Specific HRGP Motifs
2.4. Integrative Analysis Filter B: A Rigorous Set of Pollen-Specific HRGP Motifs
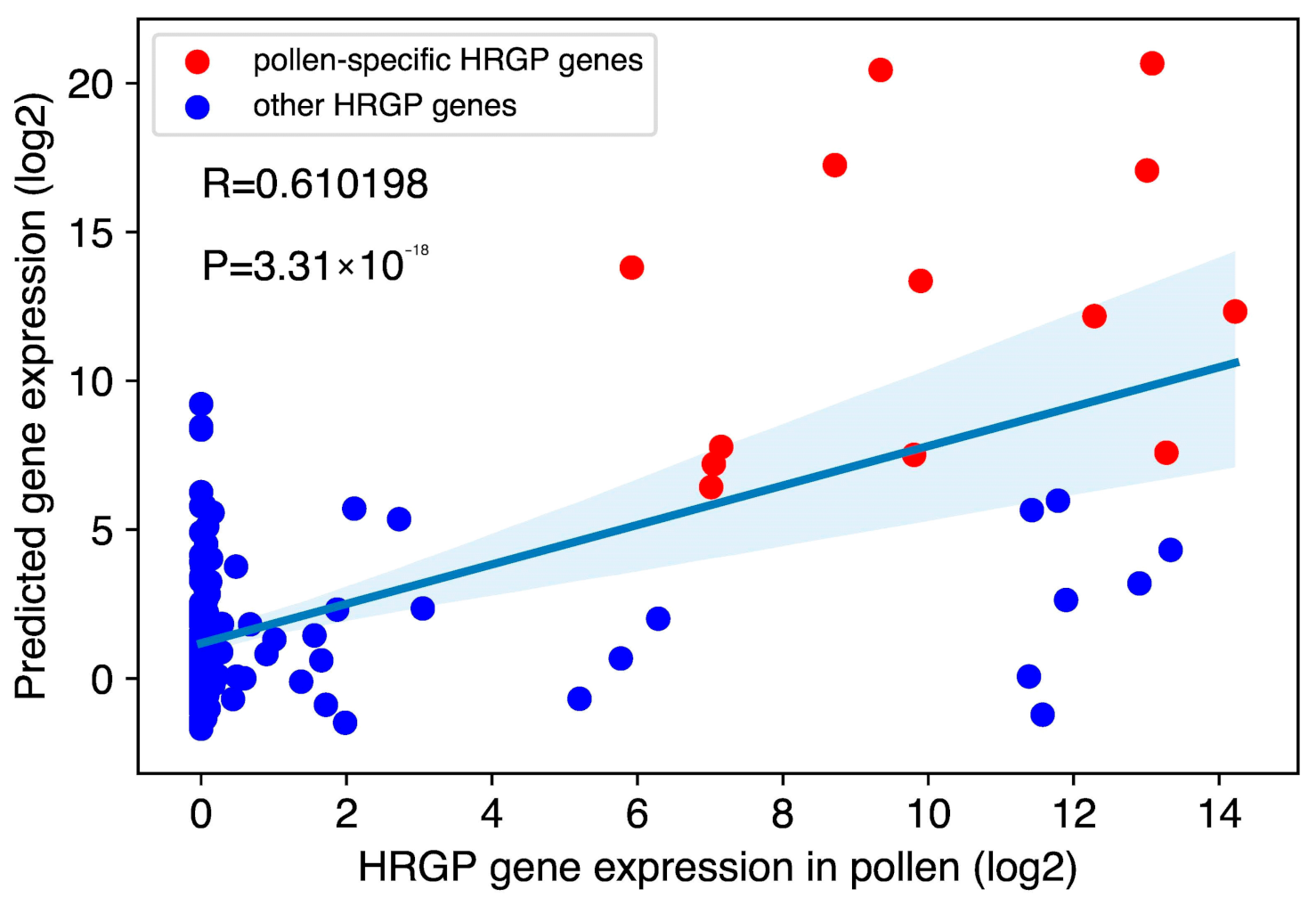
2.5. Modeling HRGP Gene Expression in Pollen
3. Discussion
4. Materials and Methods
4.1. Characterization of Pollen-Specific HRGP Genes
4.2. Promoter Retrieval and Ensemble Motif Discovery
4.3. Conservation Analysis
4.4. Machine Learning
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Zambelli, F.; Pesole, G.; Pavesi, G. Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Brief. Bioinform. 2012, 14, 225–237. [Google Scholar] [CrossRef]
- Arvey, A.; Agius, P.; Noble, W.S.; Leslie, C. Sequence and chromatin determinants of cell-type–specific transcription factor binding. Genome Res. 2012, 22, 1723–1734. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, A.; Yardimci, G.G.; Sheffield, N.C.; Crawford, G.E.; Ohler, U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012, 22, 1711–1722. [Google Scholar] [CrossRef] [Green Version]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanai, I.; Benjamin, H.; Shmoish, M.; Chalifa-Caspi, V.; Shklar, M.; Ophir, R.; Bar-Even, A.; Horn-Saban, S.; Safran, M.; Domany, E.; et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 2005, 21, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grote, A.; Li, Y.; Liu, C.; Voronin, D.; Geber, A.; Lustigman, S.; Unnasch, T.R.; Welch, L.; Ghedin, E. Prediction pipeline for discovery of regulatory motifs associated with Brugia malayi molting. PLoS Negl. Trop. Dis. 2020, 14, e0008275. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Heeringen, S.J.; Veenstra, G.J.C. GimmeMotifs: A de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics 2010, 27, 270–271. [Google Scholar] [CrossRef] [Green Version]
- Huggins, P.; Zhong, S.; Shiff, I.; Beckerman, R.; Laptenko, O.; Prives, C.; Schulz, M.H.; Simon, I.; Bar-Joseph, Z. DECOD: Fast and accurate discriminative DNA motif finding. Bioinformatics 2011, 27, 2361–2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.D.; Sumazin, P.; Zhang, M.Q. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc. Natl. Acad. Sci. USA 2005, 102, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Yu, M.; Geng, L.-L.; Zhao, J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010, 64, 482–497. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Knox, R.B.; Taylor, P.E.; Singh, M.B. Bcp1, a gene required for male fertility in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 2106–2110. [Google Scholar] [CrossRef] [Green Version]
- Coimbra, S.; Costa, M.; Jones, B.; Mendes, M.A.; Pereira, L.G. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J. Exp. Bot. 2009, 60, 3133–3142. [Google Scholar] [CrossRef] [Green Version]
- Levitin, B.; Richter, D.; Markovich, I.; Zik, M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J. 2008, 56, 351–363. [Google Scholar] [CrossRef]
- Lin, S.; Yue, X.; Miao, Y.; Yu, Y.; Dong, H.; Huang, L.; Cao, J. The distinct functions of two classical arabinogalactan proteins BcMF8 and BcMF18 during pollen wall development in Brassica campestris. Plant J. 2018, 94, 60–76. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Y.; Sae-Seaw, J.; Wang, Z.-Y. Brassinosteroid signalling. Development 2013, 140, 1615–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Wang, Q.; Chong, K.; Wang, F.; Wang, L.; Bai, M.; Jia, C. The brassinosteroid signal transduction pathway. Cell Res. 2006, 16, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P.P. A Novel RGL2-DOF6 Complex Contributes to Primary Seed Dormancy in Arabidopsis thaliana by Regulating a GATA Transcription Factor. Mol. Plant 2017, 10, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wolfe, R.; Welch, L.R.; Domozych, D.S.; Popper, Z.A.; Showalter, A.M. Bioinformatic Identification and Analysis of Extensins in the Plant Kingdom. PLoS ONE 2016, 11, e0150177. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.D.; Liu, X.; Lichtenberg, J.; Welch, L.R. Bioinformatic Identification and Analysis of Hydroxyproline-Rich Glycoproteins in Populus trichocarpa. BMC Plant Biol. 2016, 16, 229. [Google Scholar] [CrossRef] [Green Version]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [Green Version]
- Waskom, M.; Botvinnik, O.; O’Kane, D.; Hobson, P.; Lukauskas, S.; Gemperline, D.C.; Augspurger, T.; Halchenko, Y.; Cole, J.B.; Warmenhoven, J.; et al. mwaskom/seaborn: v0.8.1 (September 2017). Zenodo 2017. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, G.; Mereghetti, P.; Mauri, G.; Pesole, G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004, 32, W199–W203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Brutlag, D.L.; Liu, J.S. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001, 2001, 127–138. [Google Scholar]
- Shi, J.; Yang, W.; Chen, M.; Du, Y.; Zhang, J.; Wang, K. AMD, an Automated Motif Discovery Tool Using Stepwise Refinement of Gapped Consensuses. PLoS ONE 2011, 6, e24576. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Christopher, K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L. GADEM: A Genetic Algorithm Guided Formation of Spaced Dyads Coupled with an EM Algorithm for Motif Discovery. J. Comput. Biol. 2009, 16, 317–329. [Google Scholar] [CrossRef]
- Conlon, E.M.; Liu, X.S.; Lieb, J.D.; Liu, J.S. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 3339–3344. [Google Scholar] [CrossRef] [Green Version]
- Ao, W.; Gaudet, J.; Kent, W.J.; Muttumu, S.; Mango, S.E. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 2004, 305, 1743–1746. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Kagda, M.; Judelson, H.S. Genome-wide Prediction and Functional Validation of Promoter Motifs Regulating Gene Expression in Spore and Infection Stages of Phytophthora infestans. PLoS Pathog. 2013, 9, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]

| TAIR ID | Gene Name a | Tissue Specificity Index b | Expression in Pollen c | Level of Expression d | Reference e |
|---|---|---|---|---|---|
| AT1G10620 * | PERK11 | 0.968 | 7.003 | Extremely high | [13] |
| AT1G49270 * | PERK7 | 0.960 | 8.711 | Extremely high | [13] |
| AT4G34440 * | PERK5 | 0.948 | 7.039 | Extremely high | [13] |
| AT3G18810 * | PERK6 | 0.941 | 9.343 | Extremely high | [13] |
| AT2G24450 * | FLA3 | 0.936 | 12.284 | Extremely high | [14] |
| AT1G23540 * | PERK12 | 0.936 | 7.141 | Extremely high | [13] |
| AT4G33970 * | PEX4 | 0.909 | 9.893 | Extremely high | [15] |
| AT1G54215 | EXT32 | 0.908 | 5.898 | High | |
| AT2G18470 * | PERK4 | 0.880 | 9.803 | Extremely high | [13] |
| AT1G24520 * | AGP50 | 0.879 | 13.274 | Extremely high | [16] |
| AT3G01700 * | AGP11 | 0.872 | 13.079 | Extremely high | [17,18] |
| AT5G14380 * | AGP6 | 0.862 | 13.008 | Extremely high | [17,18] |
| AT3G57690 * | AGP23 | 0.856 | 14.220 | Extremely high | [19] |
| Motif Name | Motif Logo | Foreground Coverage | Background Coverage | Best Matched TFBS (p-Value) |
|---|---|---|---|---|
| DME_GADGAYKAS |  | 85% (11) | 19% (25) | AT3G11280, MYB-LIKE PROTEIN (3.6 × 10−4) |
| DME_GATYTKRHG |  | 85% (11) | 20% (27) | |
| DME_GRHTGDTGA |  | 85% (11) | 20% (27) | AT5G58620, TZF9 (1.3 × 10−5) |
| DME_MARKGDSRGA |  | 85% (11) | 22% (29) | |
| gimme_102_Improbizer_GCGTTATACCCGAGGATCAG | 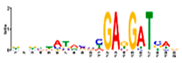 | 92% (12) | 15% (20) | |
| gimme_104_Improbizer_GTGCAACGGAGAGT | 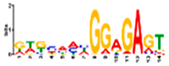 | 92% (12) | 14% (18) | |
| gimme_105_Improbizer_AACACACGTTTATTAGATGTTT | 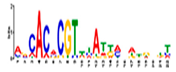 | 100% (13) | 18% (24) | AT1G75080, BZR1 (1.6 × 10−6) |
| gimme_132_MEME_3_w10 |  | 92% (12) | 11% (15) | |
| gimme_13_BioProspector_w10_3 |  | 85% (11) | 13% (17) | |
| gimme_143_MEME_4_w12 |  | 85% (11) | 12% (16) | AT5G11260, TED5 (4.7 × 10−4) |
| gimme_146_MEME_7_w12 |  | 92% (12) | 16% (21) | |
| gimme_16_BioProspector_w12_1 |  | 85% (11) | 20% (27) | |
| gimme_92_MDmodule_Motif.12.3 |  | 85% (11) | 20% (26) |
| Motif Name | Motif Logo | Foreground Coverage | Background Coverage | Best Matched TFBS (p-Value) |
|---|---|---|---|---|
| DME_ACDGWGMYA |  | 77% (10) | 10% (13) | AT4G32890, GATA9 (5 × 10−4) |
| gimme_143_MEME_4_w12 |  | 85% (11) | 12% (16) | AT5G11260, TED5 (9 × 10−4) |
| DME_ARRTCYKVRG |  | 77% (10) | 12% (16) | AT4G32890, GATA9 (1.8 × 10−3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Mullin, M.; Zhang, Y.; Drews, F.; Welch, L.R.; Showalter, A.M. Identification of Cis-Regulatory Sequences Controlling Pollen-Specific Expression of Hydroxyproline-Rich Glycoprotein Genes in Arabidopsis thaliana. Plants 2020, 9, 1751. https://doi.org/10.3390/plants9121751
Li Y, Mullin M, Zhang Y, Drews F, Welch LR, Showalter AM. Identification of Cis-Regulatory Sequences Controlling Pollen-Specific Expression of Hydroxyproline-Rich Glycoprotein Genes in Arabidopsis thaliana. Plants. 2020; 9(12):1751. https://doi.org/10.3390/plants9121751
Chicago/Turabian StyleLi, Yichao, Maxwell Mullin, Yingnan Zhang, Frank Drews, Lonnie R. Welch, and Allan M. Showalter. 2020. "Identification of Cis-Regulatory Sequences Controlling Pollen-Specific Expression of Hydroxyproline-Rich Glycoprotein Genes in Arabidopsis thaliana" Plants 9, no. 12: 1751. https://doi.org/10.3390/plants9121751





