Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane
Abstract
1. Introduction
2. Results
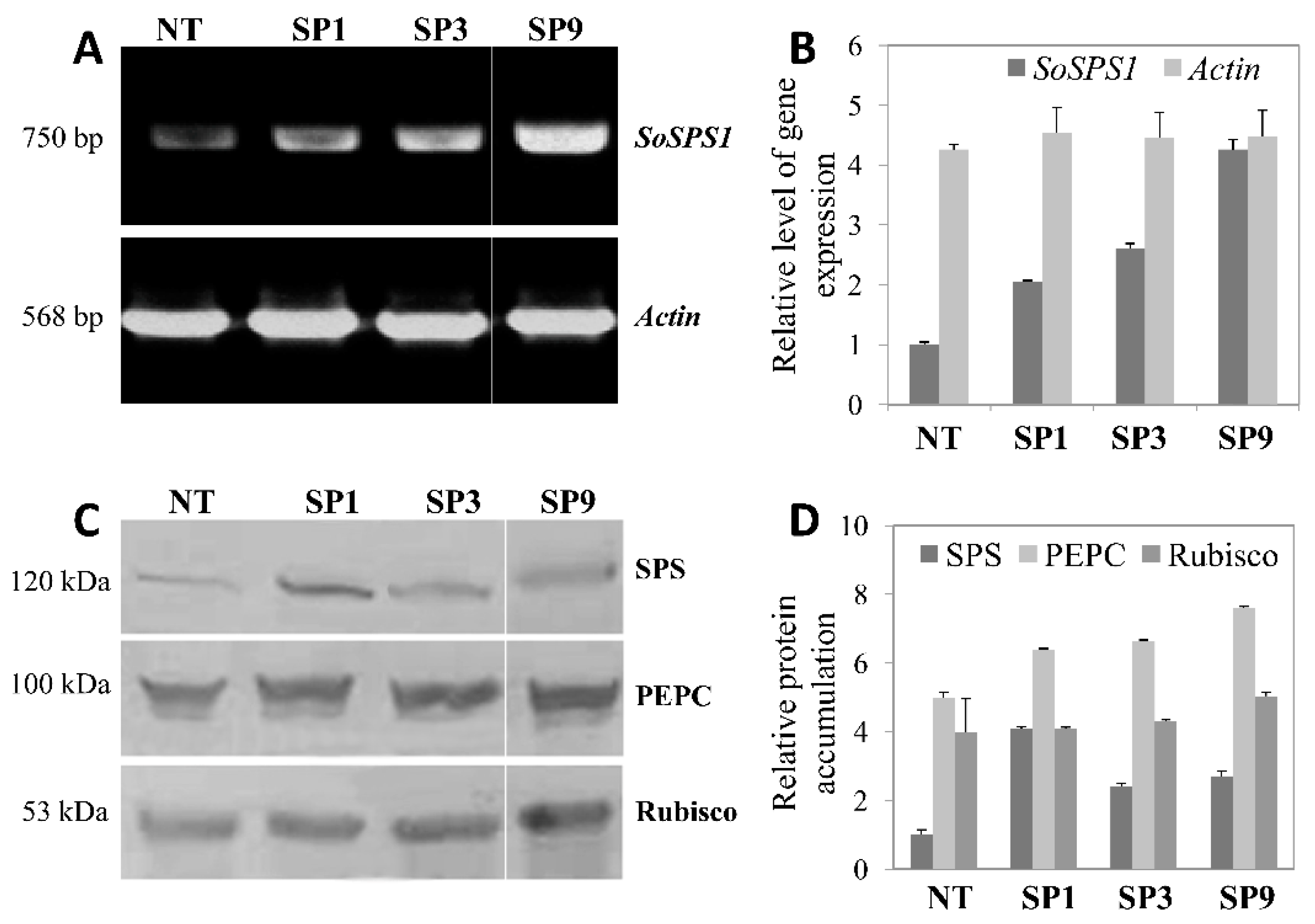
2.1. Expression of SoSPS1 Gene in Transgenic Sugarcane
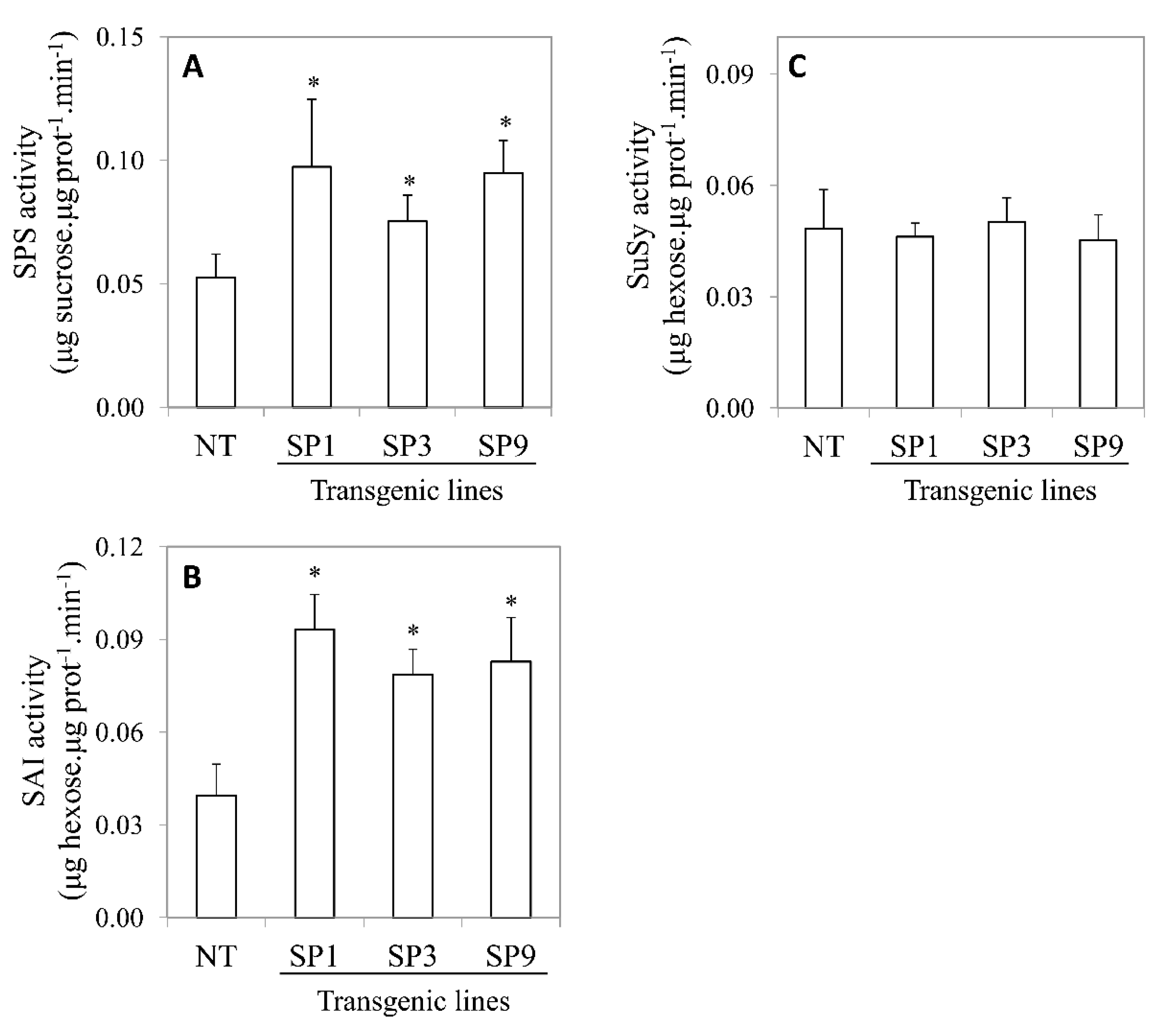
2.2. Sucrose Metabolizing Enzymes Activities
2.3. Increasing Sugar Content in the Leaves and Stalks of Transgenic Sugarcane
2.4. The Effect of SPS Overexpression on Sugarcane Growth
3. Discussion
4. Materials and Methods
4.1. Plant Transformation and Growth Condition
4.2. Genomic and Gene Expression Analysis
4.3. Protein Extraction, Enzyme Assay, and Immunoblotting
4.4. Sugar Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Sturm, A. Invertases. Primary Structures, Functions, and Roles in Plant Development and Sucrose Partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef]
- Pan, Y.Q.; Luo, H.L.; Li, Y.R. Soluble acid invertase and sucrose phosphate synthase: Key enzymes in regulating sucrose accumulation in sugarcane stalk. Sugar Tech 2009, 11, 28–33. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Komor, E.; Moore, P.H. Sucrose Accumulation in the Sugarcane Stalk Is Regulated by the Difference between the Activities of Soluble Acid Invertase and Sucrose Phosphate Synthase. Plant Physiol. 1997, 115, 609–616. [Google Scholar] [CrossRef]
- Worrell, A.C.; Bruneau, J.-M.; Summerfelt, K.; Boersig, M.; Voelker, T.A. Expression of a Maize Sucrose Phosphate Synthase in Tomato Alters Leaf Carbohydrate Partitioning. Plant Cell 2007, 3, 1121. [Google Scholar] [CrossRef]
- Park, J.Y.; Canam, T.; Kang, K.Y.; Ellis, D.D.; Mansfield, S.D. Over-expression of an Arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef]
- Sugiharto, B.; Sakakibara, H.; Sumadi; Sugiyama, T. Differential expression of two genes for sucrose-phosphate synthase in sugarcane: Molecular cloning of the cDNAs and comparative analysis of gene expression. Plant Cell Physiol. 1997, 38, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Falter, C.; Voigt, C.A. Improving biomass production and saccharification in Brachypodium distachyon through overexpression of a sucrose-phosphate synthase from sugarcane. J. Plant Biochem. Biotechnol. 2016, 25, 311–318. [Google Scholar] [CrossRef][Green Version]
- Galtier, N.; Foyer, C.H.; Huber, J.; Voelker, T.A.; Huber, S.C. Effects of Elevated Sucrose-Phosphate Synthase Activity on Photosynthesis, Assimilate Partitioning, and Growth in Tomato (Lycopersicon esculentum var UC82B). Plant Physiol. 1993, 101, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Signora, L.; Galtier, N.; Skot, L.; Lucas, H.; Foyer, C.H. Over-expression of sucrose phosphate synthase in Arabidopsis thaliana results in increased foliar sucrose/starch ratios and favours decreased foliar carbohydrate accumulation in plants after prolonged growth with CO2 enrichment. J. Exp. Bot. 1998, 49, 669–680. [Google Scholar] [CrossRef]
- Nguyen-Quoc, B.; N’Tchobo, H.; Foyer, C.H.; Yelle, S. Overexpression of sucrose phosphate synthase increases sucrose unloading in transformed tomato fruit. J. Exp. Bot. 1999, 50, 785–791. [Google Scholar] [CrossRef]
- Baxter, C.J.; Foyer, C.H.; Turner, J.; Rolfe, S.A.; Quick, W.P. Elevated sucrose-phosphate synthase activity in transgenic tobacco sustains photosynthesis in older leaves and alters development. J. Exp. Bot. 2003, 54, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Maloney, V.J.; Park, J.Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase and sucrose phosphate phosphatase interact in planta and promote plant growth and biomass accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef]
- Lobo, A.K.M.; de Oliveira Martins, M.; Lima Neto, M.C.; Machado, E.C.; Ribeiro, R.V.; Silveira, J.A.G. Exogenous sucrose supply changes sugar metabolism and reduces photosynthesis of sugarcane through the down-regulation of Rubisco abundance and activity. J. Plant Physiol. 2015, 179, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Jain, R.; Solomon, S. Complexities of invertases controlling sucrose accumulation and retention in sugarcane. Curr. Sci. 2012, 102, 857–866. [Google Scholar]
- Wang, L.; Li, X.-R.; Lian, H.; Ni, D.-A.; He, Y.-K.; Chen, X.-Y.; Ruan, Y.-L. Evidence That High Activity of Vacuolar Invertase Is Required for Cotton Fiber and Arabidopsis Root Elongation through Osmotic Dependent and Independent Pathways, Respectively. Plant Physiol. 2010, 154, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Feng, J.; Qin, Q.; Huang, J. Overexpression of a loquat (Eriobotrya japonica Lindl.) vacuolar invertase affects sucrose levels and growth. Plant Cell Tissue Organ Cult. 2015, 123, 99–108. [Google Scholar] [CrossRef]
- Jain, R.; Singh, S.P.; Singh, A.; Singh, S.; Kishor, R.; Singh, R.K.; Chandra, A.; Solomon, S. Soluble Acid Invertase (SAI) Activity and Gene Expression Controlling Sugar Composition in Sugarcane. Sugar Tech 2017, 19, 669–674. [Google Scholar] [CrossRef]
- Shivalingamurthy, S.G.; Anangi, R.; Kalaipandian, S.; Glassop, D.; King, G.F.; Rae, A.L. Identification and Functional Characterization of Sugarcane Invertase Inhibitor (ShINH1): A Potential Candidate for Reducing Pre- and Post-harvest Loss of Sucrose in Sugarcane. Front. Plant Sci. 2018, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Kossmann, J.; Botha, F.C.; Groenewald, J.-H. Reduced neutral invertase activity in the culm tissues of transgenic sugarcane plants results in a decrease in respiration and sucrose cycling and an increase in the sucrose to hexose ratio. Funct. Plant Biol. 2010, 37, 22. [Google Scholar] [CrossRef]
- Fan, J.; Wang, H.; Li, X.; Sui, X.; Zhang, Z. Down-Regulating Cucumber Sucrose Synthase 4 (CsSUS4) Suppresses the Growth and Development of Flowers and Fruits. Plant Cell Physiol. 2019, 60, 931. [Google Scholar] [CrossRef] [PubMed]
- Gebril, S.; Seger, M.; Villanueva, F.M.; Ortega, J.L.; Bagga, S.; Sengupta-Gopalan, C. Transgenic alfalfa (Medicago sativa) with increased sucrose phosphate synthase activity shows enhanced growth when grown under N2-fixing conditions. Planta 2015, 242, 1009–1024. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Hirotsu, N.; Kashiwagi, T.; Madoka, Y.; Nagasuga, K.; Ono, K.; Ohsugi, R. Overexpression of a Maize SPS Gene Improves Yield Characters of Potato under Field Conditions. Plant Prod. Sci. 2008, 11, 104–107. [Google Scholar] [CrossRef]
- Seger, M.; Gebril, S.; Tabilona, J.; Peel, A.; Sengupta-Gopalan, C. Impact of concurrent overexpression of cytosolic glutamine synthetase (GS1) and sucrose phosphate synthase (SPS) on growth and development in transgenic tobacco. Planta 2015, 241, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Lunn, J.; Usadel, B. Arabidopsis and primary photosynthetic metabolism-more than the icing on the cake. Plant J. 2010, 61, 1067–1091. [Google Scholar] [CrossRef]
- McCormick, A.J.; Watt, D.A.; Cramer, M.D. Supply and demand: Sink regulation of sugar accumulation in sugarcane. J. Exp. Bot. 2009, 60, 357–364. [Google Scholar] [CrossRef]
- Inman-Bamber, N.G.; Jackson, P.A.; Hewitt, M. Sucrose accumulation in sugarcane stalks does not limit photosynthesis and biomass production. Crop Pasture Sci. 2011, 62, 848. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Smeekens, S.; Hellmann, H.A. Sugar Sensing and Signaling in Plants. Plant Cell 2014, 14, S185–S205. [Google Scholar] [CrossRef]
- Eveland, A.L.; Jackson, D.P. Sugars, signalling, and plant development. J. Exp. Bot. 2012, 63, 3367–3377. [Google Scholar] [CrossRef]
- De Avila Silva, L.; Condori-Apfata, J.A.; de Almeida Costa, P.M.; Martino, P.B.; Tavares, A.C.A.; Marcelino, M.M.; Raimundi, S.C.J.; de Toledo Picoli, E.A.; Araujo, W.L.; Zsogon, A.; et al. Source Strength Modulates Fruit Set by Starch Turnover and Export of Both Sucrose and Amino Acids in Pepper. Plant Cell Physiol. 2019, 60, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Rosche, E.; Blackmore, D.; Tegeder, M.; Richardson, T.; Schroeder, H.; Higgins, T.J.V.; Frommer, W.B.; Offler, C.E.; Patrick, J.W. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 2002, 30, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Leggewie, G.; Kolbe, A.; Lemoine, R.; Roessner, U.; Lytovchenko, A.; Zuther, E.; Kehr, J.; Frommer, W.B.; Riesmeier, J.W.; Willmitzer, L.; et al. Overexpression of the sucrose transporter So SUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta 2003, 217, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wen, S.; Xiao, S.; Lu, B.; Ma, M.; Bie, Z. Overexpression of the tonoplast sugar transporter CmTST2 in melon fruit increases sugar accumulation. J. Exp. Bot. 2018, 69, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 204–207. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stalk sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef]
- Sugiyama, A.; Saida, Y.; Yoshimizu, M.; Takanashi, K.; Sosso, D.; Frommer, W.B.; Yazaki, K. Molecular Characterization of LjSWEET3, a Sugar Transporter in Nodules of Lotus japonicus. Plant Cell Physiol. 2017, 58, 298–306. [Google Scholar]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Sawitri, W.D.; Narita, H.; Ishizaka-Ikeda, E.; Sugiharto, B.; Hase, T.; Nakagawa, A. Purification and characterization of recombinant sugarcane sucrose phosphate synthase expressed in E. coli and insect Sf9 cells: An importance of the N-Terminal domain for an allosteric regulatory property. J. Biochem. 2016, 159, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Apriasti, R.; Widyaningrum, S.; Hidayati, W.N.; Sawitri, W.D.; Darsono, N.; Hase, T.; Sugiharto, B. Full sequence of the coat protein gene is required for the induction of pathogen-derived resistance against sugarcane mosaic virus in transgenic sugarcane. Mol. Biol. Rep. 2018, 45, 2749–2758. [Google Scholar] [CrossRef] [PubMed]
- Gerber, L.; Zhang, B.; Roach, M.; Rende, U.; Gorzsás, A.; Kumar, M.; Burgert, I.; Niittylä, T.; Sundberg, B. Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytol. 2014, 203, 1220–1230. [Google Scholar] [CrossRef]
- Garriga, M.; Almaraz, M.; Marchiaro, A. Determination of reducing sugars in extracts of Undaria pinnatifida (harvey) algae by UV-visible spectrophotometry (DNS method). Actas De Ing. 2017, 3, 173–179. [Google Scholar]
- Sugiharto, B.; Ermawati, N.; Mori, H.; Aoki, K.; Yonekura-Sakakibara, K.; Yamaya, T.; Sugiyama, T.; Sakakibara, H. Identification and characterization of a gene encoding drought-inducible protein localizing in the bundle sheath cell of sugarcane. Plant Cell Physiol. 2002, 43, 350–354. [Google Scholar] [CrossRef][Green Version]
| Lines | Leaf Tissue | Stalk Tissue | ||||
|---|---|---|---|---|---|---|
| Sucrose (mg/g FW) | Fructose (mg/g FW) | Glucose (mg/g FW) | Sucrose (mg/g FW) | Fructose (mg/g FW) | Glucose (mg/g FW) | |
| NT | 2.27 ± 0.10 c | 0.35 ± 0.17 b | 0.18 ± 0.06 b | 71.07 ± 3.30 b | 2.33 ± 0.31 b | 3.23 ± 1.02 c |
| SP1 | 3.59 ± 0.04 b | 3.34 ± 0.39 a | 2.21 ± 0.58 a | 80.40 ± 8.32 b | 2.87 ± 0.46 ab | 4.40 ± 1.15 bc |
| SP3 | 5.51 ± 0.24 a | 3.38 ± 0.58 a | 1.52 ± 0.64 ab | 94.23 ± 3.34 a | 3.62 ± 0.20 a | 6.25 ± 1.06 a |
| SP9 | 3.02 ± 0.34 b | 2.47 ± 0.08 a | 1.30 ± 0.83 ab | 98.52 ± 5.55 a | 3.31 ± 0.26 ab | 4.86 ± 0.30 ab |
| Lines | Plant Height (cm) | Stalk Diameter (cm) | Stalk Number | Stalk Weight per Pot (g) |
|---|---|---|---|---|
| NT | 99.67 ± 3.67 b | 2.24 ± 0.03 b | 9.00 ± 1.00 b | 3537.90 ± 680 b |
| SP1 | 115.33 ± 6.00 a | 2.38 ± 0.03 a | 10.67 ± 1.53 ab | 4193.06 ± 600 ab |
| SP3 | 112.78 ± 5.01 a | 2.24 ± 0.04 b | 12.67 ± 0.58 a | 4979.26 ± 226 a |
| SP9 | 124.33 ± 3.93 a | 2.18 ± 0.05 b | 11.67 ± 0.58 a | 4586.16 ± 226 ab |
| Primer Names | Sequence (5′–3′) | Product (bp) | Target Genes |
|---|---|---|---|
| F1 | TGAATGAACTGCAGGACGAG | 550 | npt II |
| R1 | AGCCAACGTATGTCCTGAT | 550 | npt II |
| F2 | TGAAGGACACACCGGCAGATG | 750 | SoSPS1 |
| R2 | CTTTGATGAGGAAGGCGAAGC | 750 | SoSPS1 |
| F3 | GCAACTGGGATGACATGGAG | 568 | Actin |
| R3 | ATGGCTGGAAGAGGACCTCAG | 568 | Actin |
| F4 | TGCACTAGTCGCCCTTCCCA | 3425 | SoSPS1 |
| R4 | TCCACTAGTAACGGCCGCCA | 3425 | SoSPS1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anur, R.M.; Mufithah, N.; Sawitri, W.D.; Sakakibara, H.; Sugiharto, B. Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane. Plants 2020, 9, 200. https://doi.org/10.3390/plants9020200
Anur RM, Mufithah N, Sawitri WD, Sakakibara H, Sugiharto B. Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane. Plants. 2020; 9(2):200. https://doi.org/10.3390/plants9020200
Chicago/Turabian StyleAnur, Risky Mulana, Nurul Mufithah, Widhi Dyah Sawitri, Hitoshi Sakakibara, and Bambang Sugiharto. 2020. "Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane" Plants 9, no. 2: 200. https://doi.org/10.3390/plants9020200
APA StyleAnur, R. M., Mufithah, N., Sawitri, W. D., Sakakibara, H., & Sugiharto, B. (2020). Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane. Plants, 9(2), 200. https://doi.org/10.3390/plants9020200





