Reducing Cadmium Accumulation in Plants: Structure–Function Relations and Tissue-Specific Operation of Transporters in the Spotlight
Abstract
:1. Cd Toxicity as an Issue
1.1. Cadmium in Soils
1.2. Cadmium Toxicity: Health Implications
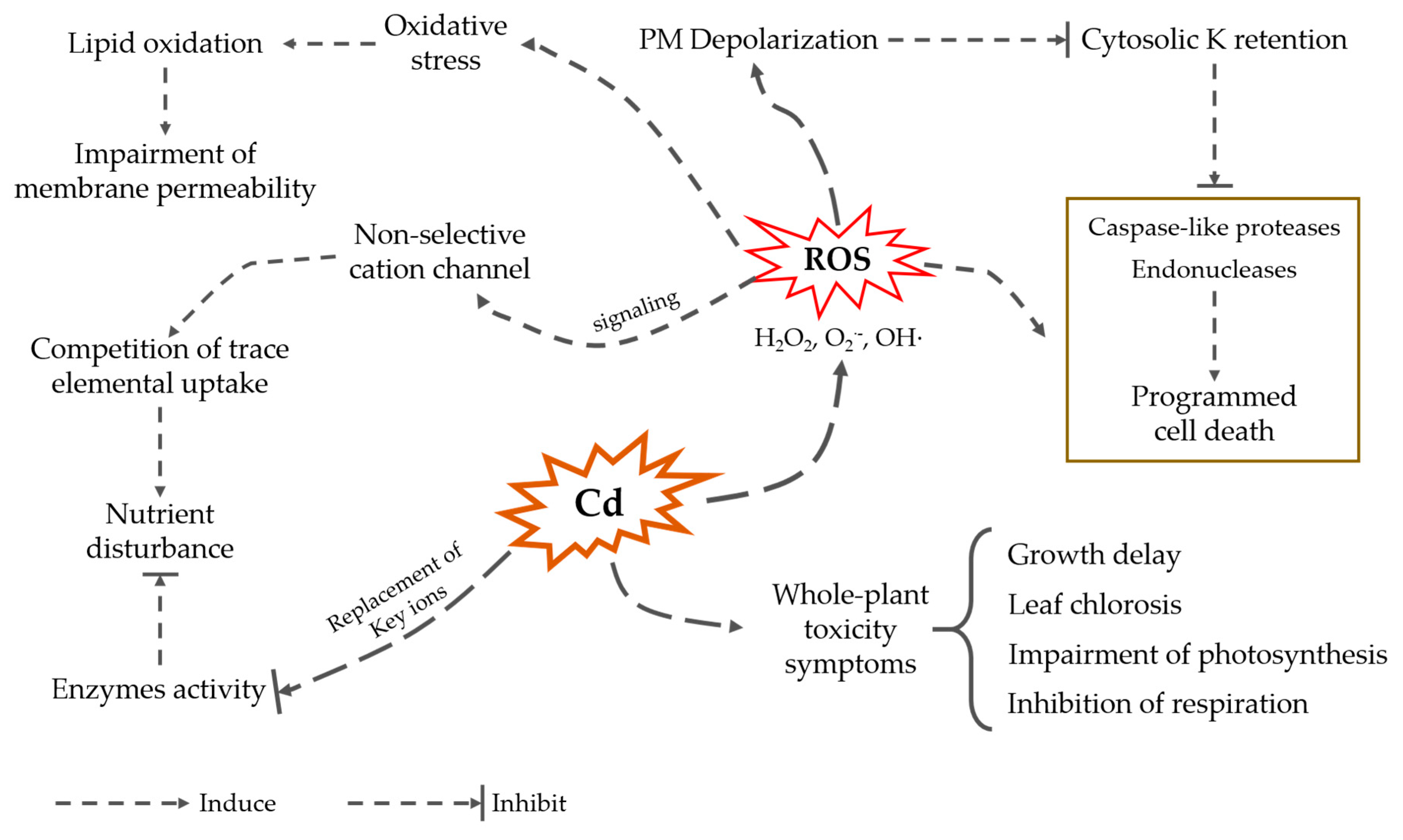
1.3. Cadmium Impact on Plant Growth and Performance
2. Current Trends in Breeding Programs
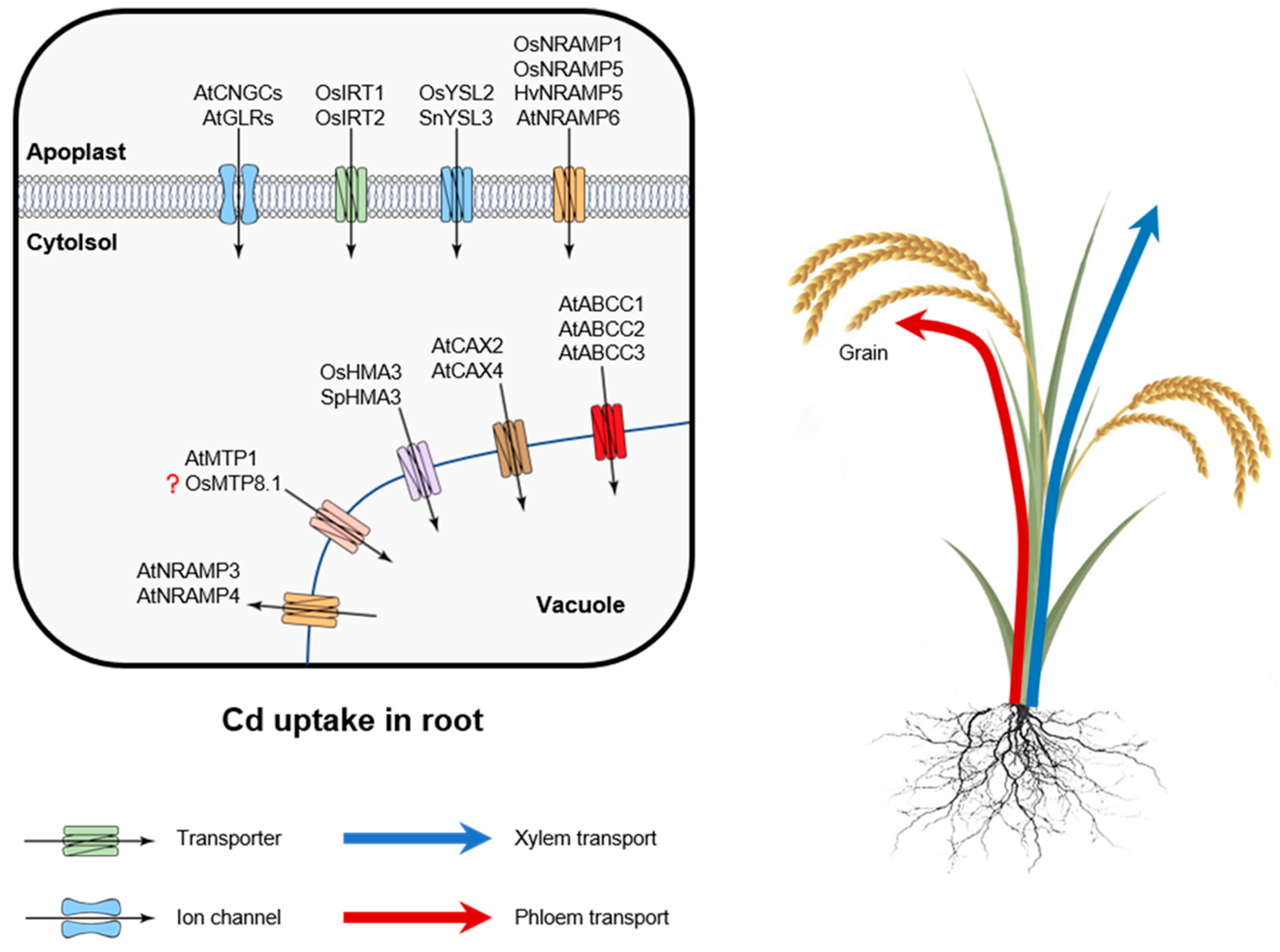
3. Molecular Mechanisms of Cd Uptake and Transport
3.1. Cd Uptake by Roots
3.2. Long-Distant Cd Transport
4. Tissue Tolerance Mechanisms
5. Conclusions and the Way Forward
Author Contributions
Funding
Conflicts of Interest
References
- Panos, P.; Marc, V.L.; Yusuf, Y.; Luca, M. Contaminated sites in Europe: Review of the current situation based on data collected through a European network. J. Environ. Public Health 2013, 2013, 158764. [Google Scholar]
- Alloway, B.J. Heavy Metals in Soils: Trace Metals and Metalloids in Soils and Their Bioavailability; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Cook, M.E.; Morrow, H. Anthropogenic sources of cadmium in Canada. In National Workshop on Cadmium Transport into Plants; Canadian Network of Toxicology Centres: Ottawa, ON, Canada, 1995. [Google Scholar]
- Hayat, M.T.; Nauman, M.; Nazir, N.; Ali, S.; Bangash, N. Environmental hazards of cadmium: Past, present, and future. In Cadmium Toxicity and Tolerance in Plants. From Physiology to Remediation; Hasanuzzaman, M., Prasad, M.N.V., Fujita, M., Eds.; Academic Press: London, UK, 2019; pp. 163–183. [Google Scholar]
- World Health Organisation (WHO). Environmental Health Criteria 134—Cadmium International Programme on Chemical Safety (IPCS) Monograph; World Health Organization: Geneva, Switzerland, 1992. [Google Scholar]
- Xue, D.W.; Jiang, H.; Deng, X.X.; Zhang, X.Q.; Wang, H.; Xu, X.B.; Hu, J.; Zeng, D.; Guo, L.; Qian, Q. Comparative proteomic analysis provides new insights into cadmium accumulation in rice grain under cadmium stress. J. Hazard. Mater. 2014, 280, 269–278. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [Green Version]
- Zu, Y.; Li, Y.; Chen, J.; Chen, H.Y.; Li, Q.; Schratz, C. Hyper accumulation of Pb, Zn and Cd in herbaceous grown on lead-zinc mining area in Yunnan, China. Environ. Int. 2005, 1, 755–762. [Google Scholar]
- Sanita, D.T.L.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Zhao, K.L.; Fu, W.J.; Ye, Z.Q.; Zhang, C.S. Contamination and spatial variation of heavy metals in the soil-rice system in Nanxun County, Southeastern China. J. Environ. Res. Public Health 2015, 12, 1577–1594. [Google Scholar] [CrossRef] [PubMed]
- Bauddh, K.; Singh, R.P. Growth, tolerance efficiency and phytoremediation potential of Ricinus communis (L.) and Brassica juncea (L.) in salinity and drought affected cadmium contaminated soil. Ecotoxicol. Environ. Saf. 2012, 85, 13–22. [Google Scholar] [CrossRef]
- Shi, G.; Xia, S.; Ye, J.; Huang, Y.; Liu, C.; Zhang, Z. PEG-simulated drought stress decreases cadmium accumulation in castor bean by altering root morphology. Environ. Exp. Bot. 2015, 111, 127–134. [Google Scholar] [CrossRef]
- Demeyer, A.; Nkana, J.V.; Verloo, M.G. Characteristics of wood ash and influence on soil properties and nutrient uptake: An overview. Bioresour. Technol. 2001, 77, 287–295. [Google Scholar] [CrossRef]
- Kabata-Pendias, A. Trace Elements in Soils and Plants, 4th ed.; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Smith, S.R. A critical review of the bioavailability and impacts of heavy metals in municipal solid waste composts compared to sewage sludge. Environ. Int. 2009, 35, 142–156. [Google Scholar] [CrossRef]
- De Oliveira, V.H.; Tibbett, M. Cd and Zn interactions and toxicity in ectomycorrhizal basidiomycetes in axenic culture. PeerJ 2018, 6, e4478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, J.L.; Rønn, R.; Ekelund, F. Toxicity of cadmium and zinc to small soil protists. Environ. Pollut. 2018, 242, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 2016, 1, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, T.; Plusquin, M.; Hogervorst, J.; Roels, H.A.; Celis, H.; Thijs, L.; Vangronsveld, J.; Van Hecke, E.; Staessen, J.A. Environmental exposure to cadmium and risk of cancer: A prospective populationbased study. Lancet Oncol. 2006, 7, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Tsukahara, T.; Ezaki, T.; Moriguchi, J.; Furuki, K.; Shimbo, S.; Matsuda-Inoguchi, N.; Ikeda, M. Rice as the most influential source of cadmium intake among general Japanese population. Sci. Total Environ. 2003, 305, 41–51. [Google Scholar] [CrossRef]
- Inaba, T.; Kobayashi, E.; Suwazono, Y.; Uetani, M.; Oishi, M.; Nakagawa, H.; Nogawa, K. Estimation of cumulative cadmium intake causing Itai-itai disease. Toxicol. Lett. 2005, 159, 192–201. [Google Scholar] [CrossRef]
- Kara, H.; Karatas, F.; Canatan, H.; Servi, K. Effects of exogenous metallothionein on acute cadmium toxicity in rats. Biol. Trace Elem. Res. 2005, 104, 223. [Google Scholar] [CrossRef]
- Dudley, R.E.; Svoboda, D.J.; Klaassen, C.D. Acute exposure to cadmium causes severe liver injury in rats. Toxicol. Appl. Pharm. 1982, 65, 302–313. [Google Scholar] [CrossRef]
- Touceda-González, M.; Brader, G.; Antonielli, L.; Ravindran, V.B.; Waldner, G.; Friesl-Hanl, W.; Corretto, E.; Campisano, A.; Pancher, M.; Angela, S. Combined amendment of immobilizers and the plant growth-promoting strain Burkholderia phytofirmans PsJN favours plant growth and reduces heavy metal uptake. Soil Biol. Biochem. 2015, 91, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Van Assche, F.J. A Stepwise Model to Quantify the Relative Contribution of Different Environmental Sources to Human Cadmium Exposure. In Proceedings of the 8th International Nickel-Cadmium Battery Conference, Prague, Czech Republic, 21–22 September 1998. [Google Scholar]
- Clemens, S.; Aarts, M.G.M.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef]
- Uraguchi, S.; Fujiwara, T. Rice breaks ground for cadmium free cereals. Curr. Opin. Plant Biol. 2013, 16, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, X.N.; Ding, C.; Wu, L. The dynamic simulation of rice growth parameters under cadmium stress with the assimilation of multi-period spectral indices and crop model. Field Crops Res. 2015, 183, 225–234. [Google Scholar] [CrossRef]
- Song, W.E.; Chen, S.B.; Liu, J.F.; Chen, L.; Song, N.N.; Li, N.; Liu, B.V. Variation of Cd concentration in various rice cultivars and derivation of cadmium toxicity thresholds for paddy soil by species-sensitivity distribution. J. Integr. Agric. 2015, 14, 1845–1854. [Google Scholar] [CrossRef]
- Chen, H.; Yang, X.; Wang, P.; Wang, Z.; Li, M.; Zhao, F.J. Dietary cadmium intake from rice and vegetables and potential health risk: A case study in Xiangtan, southern China. Sci. Total Environ. 2018, 639, 271–277. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Mao, W.; Sui, H.; Yong, L.; Yang, D.; Jiang, D.; Zhang, L.; Gong, Y. Dietary cadmium exposure assessment among the Chinese population. PLoS ONE 2017, 12, e0177978. [Google Scholar] [CrossRef] [Green Version]
- Baldantoni, D.; Morra, L.; Zaccardelli, M.; Alfani, A. Cadmium accumulation in leaves of leafy vegetables. Ecotoxicol. Environ. Saf. 2016, 123, 89–94. [Google Scholar] [CrossRef]
- Wang, J.; Yu, N.; Mu, G.; Shinwari, K.I.; Shen, Z.; Zheng, L. Screening for Cd-safe cultivars of Chinese cabbage and a preliminary study on the mechanisms of Cd accumulation. Int. J. Environ. Res. Public Health 2017, 14, 395. [Google Scholar] [CrossRef] [Green Version]
- FAO Food and Agriculture Organization. ProdStat. Core Production Data Base, Core Production Data Base, Electronic Resource. 2012. Available online: http://faostat.fao.org/ (accessed on 11 October 2019).
- Mohamed, A.A.; Castagna, A.; Ranieri, A.; Sanita di Toppi, L. Cadmium tolerance in Brassica juncea roots and shoots is affected by antioxidant status and phytochelatin biosynthesis. Plant Physiol. Biochem. 2012, 57, 15–22. [Google Scholar] [CrossRef]
- Navarro-Leon, E.; Oviedo-Silva, J.; Ruiz, J.M.; Blasco, B. Possible role of HMA4a TILLING mutants of Brassica rapa in cadmium. Ecotoxicol. Environ. Saf. 2019, 180, 88–94. [Google Scholar] [CrossRef]
- Hassan, M.J.; Zhang, G.; Zhu, Z. Influence of cadmium toxicity on plant growth and nitrogen uptake in rice as affected by nitrogen form. J. Plant Nutr. 2008, 31, 251–262. [Google Scholar] [CrossRef]
- Alcantara, E.; Romera, F.J.; Canete, M.; De La Guardia, M.D. Effects of heavy metals on both induction and function of root Fe(III) reductase in Fe-deficient cucumber (Cucumis sativus L.) plants. J. Exp. Bot. 1994, 45, 1893–1898. [Google Scholar] [CrossRef]
- Sharma, V.; Pant, D. Structural basis for expanding the application of bioligand in metal bioremediation: A review. Bioresour. Technol. 2018, 252, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Cailliatte, R.; Lapeyre, B.; Briat, J.F.; Mari, S.; Curie, C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem. J. 2009, 422, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Tan, J.; Zhang, Y.; Liang, S.; Xiang, S.; Wang, H.; Chai, T. Isolation and characterization of a novel cadmium-regulated Yellow Stripe-Like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2017, 36, 281–296. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.L.; Yin, X.B.; Banuelos, G.S.; Lin, Z.Q.; Liu, Y.; Li, M.; Yuan, L.X. Indications of selenium protection against cadmium and lead toxicity in oilseed rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1875. [Google Scholar] [CrossRef]
- Lindberg, S.; Landberg, T.; Greger, M. A new method to detect cadmium uptake in protoplasts. Planta 2004, 219, 526–532. [Google Scholar] [CrossRef]
- Li, D.D.; Zhou, D.M. Toxicity and subcellular distribution of cadmium in wheat as affected by dissolved organic acids. J. Environ. Sci. 2012, 24, 903–911. [Google Scholar] [CrossRef]
- Jasinski, M.; Sudre, D.; Schansker, G.; Schellenberg, M.; Constant, S.; Martinoia, E. Atosa1, a member of the abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 2008, 147, 719–731. [Google Scholar] [CrossRef] [Green Version]
- Bortner, C.D.; Hughes, F.M.; Cidlowski, J.A. A primary role for K+ and Na+ efflux in the activation of apoptosis. J. Biol. Chem. 1977, 272, 32436–32442. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V.; Cuin, T.A.; Svistunenko, D.; Smith, S.J.; Miller, A.J.; Shabala, S.; Sokolik, A.; Yurin, V. Arabidopsis root K+ efflux conductance by hydroxyl radicals: Single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 2010, 123, 1468–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabala, S. Physiological and cellular aspects of phytotoxicity tolerance in plants: The role of membrane transporters and implications for crop breeding for waterlogging tolerance. New Phytol. 2007, 190, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Millán, A.F.; Sagardoy, R.; Solanas, M.; Abadía, A.; Abadía, J. Cadmium toxicity in tomato (Lycopersicon esculentum) plants grown in hydroponics. Environ. Exp. Bot. 2009, 65, 376–385. [Google Scholar] [CrossRef]
- Tian, S.; Lu, L.; Labavitch, J.; Yang, X.; He, Z.; Hu, H.; Sarangi, R.; Newville, M.; Commisso, J.; Brown, P. Cellular sequestration of cadmium in the hyperaccumulator plant species Sedum alfredii. Plant Physiol. 2011, 157, 1914–1925. [Google Scholar] [CrossRef] [Green Version]
- Ueno, D.; Kono, I.; Yokosho, K.; Ando, T.; Yano, M.; Ma, J.F. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol. 2009, 182, 644–653. [Google Scholar] [CrossRef]
- Ueno, D.; Koyama, E.; Kono, I.; Ando, T.; Yano, M.; Ma, J.F. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in Rice. Plant Cell Physiol. 2009, 50, 2223–2233. [Google Scholar] [CrossRef] [Green Version]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Wang, P.; Wang, P.; Yang, M.; Lian, X.; Tang, Z.; Huang, C.; Salt, D.; Zhao, F.J. A loss-of-function allele of associated with high cadmium accumulation in shoots and grain of rice cultivars. Plant Cell Environ. 2016, 39, 1941–1954. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; Sheng, Z.; Li, Q.; Chen, W.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Wang, J.; Tang, S.; et al. Identification of QTLs associated with cadmium concentration in rice grains. J. Integr. Agric. 2018, 17, 1563–1573. [Google Scholar] [CrossRef] [Green Version]
- Kuramata, M.; Masuya, S.; Takahashi, Y.; Kitagawa, E.; Inoue, C.; Ishikawa, S.; Youssefian, S.; Kusano, T. Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol. 2009, 50, 106–117. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.X.; Yamaji, N.; Ma, J.F. A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 2013, 76, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, M.; Li, Y.; Tian, J.; Zhang, Y.; Liang, L.; Liu, Z.; Chen, K.; Li, Y.; Lv, K.; et al. Comprehensive analysis of variation of cadmium accumulation in rice and detection of a new weak allele of OsHMA3. J. Exp. Bot. 2019, 70, 6389–6400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Gao, Z.; Shang, L.; Yang, C.; Ruan, B.; Zeng, D.; Guo, L.; Zhao, F.; Huang, C.; Qian, Q. Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between indica and japonica rice. J. Integr. Plant Biol. 2009, 1672–9072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Chen, S.; Chen, M.; Zheng, G.; Peng, Y.; Shi, X.; Qin, P.; Xu, X.; Teng, S. Association study reveals genetic loci responsible for arsenic, cadmium and lead accumulation in rice grain in contaminated farmlands. Front. Plant Sci. 2009, 10, 61. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing b-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wang, R.; Ju, Q.; Li, W.; Tran, L.P.; Xu, J. The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 2019, 180, 529–542. [Google Scholar] [CrossRef]
- Benitez, E.R.; Hajika, M.; Yamada, T.; Takahashi, K.; Oki, N.; Yamada, N.; Nakamura, T.; Kanamaru, K. A Major QTL controlling seed cadmium accumulation in soybean. Crop Sci. 2010, 50, 1728–1734. [Google Scholar] [CrossRef]
- Vollmann, J.; Lošák, T.; Pachner, M.; Watanabe, D.; Musilová, L.; Hlušek, J. Soybean cadmium concentration: Validation of a QTL affecting seed cadmium accumulation for improved food safety. Euphytica 2014, 203, 177–184. [Google Scholar] [CrossRef]
- Clemens, S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 2006, 88, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Hou, H.; Zhao, L.; Sun, Z.J.; Lu, Y.F.; Li, H. Mitigation of Cd accumulation in rice from Cd-contaminated paddy soil by foliar dressing of S and P. Sci. Total Environ. 2019, 690, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, F.; Liu, S.; Du, Y.; Li, F.R.; Du, R.Y.; Wen, D.; Zhao, J. Comparative responses to silicon and selenium in relation to cadmium uptake, compartmentation in roots, and xylem transport in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilis) under cadmium stress. J. Exp. Bot. 2016, 131, 173–180. [Google Scholar] [CrossRef]
- Li, Y.; Liang, X.F.; Huang, Q.Q.; Xu, Y.M.; Yang, F. Inhibition of Cd accumulation in grains of wheat and rice under rotation mode using composite silicate amendment. RSC Adv. 2019, 61, 35539–35548. [Google Scholar] [CrossRef] [Green Version]
- Lux, A.; Vaculik, M.; Martinka, M.; Liskova, D.; Kulkarni, M.G.; Stirk, W.A.; Van, S. Cadmium induces hypodermal periderm formation in the roots of the monocotyledonous medicinal plant Merwilla plumbea. Ann. Bot. 2011, 107, 285–292. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [Green Version]
- Uraguchi, S.; Kamiya, T.; Sakamoto, T.; Kasai, K.; Sato, Y.; Nagamura, Y.; Yoshida, A.; Kyozuka, J.; Ishikawa, S.; Fujiwara, T. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. USA 2011, 108, 20959–20964. [Google Scholar] [CrossRef] [Green Version]
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.F.; Walker, E.L. Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 2001, 409, 346–349. [Google Scholar] [CrossRef]
- Murata, Y.; Ma, J.F.; Yamaji, N.; Ueno, D.; Nomoto, K.; Iwashita, T. A specific transporter for iron (III)–phytosiderophore in barley roots. Plant J. 2006, 46, 563–572. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Ogawa, I.; Nakanishi, H.; Ishimaru, Y.; Takahashi, M.; Mori, S.; Nishizawa, N.K. Iron deficiency enhanced Cd uptake and translocation by Fe2+ transporters, OsIRT1 and OsIRT2, in rice. Plant Cell Physiol. 2006, 47, S231. [Google Scholar]
- Yoshihara, T.; Hodoshima, H.; Miyano, Y.; Shoji, K.; Shimada, H.; Goto, F. Cadmium inducible Fe deficiency responses observed from macro and molecular views in tobacco plants. Plant Cell Rep. 2006, 25, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Lanquar, V.; Lelièvre, F.; Barbier-Brygoo, H.; Thomine, S. Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci. 2004, 50, 1141–1150. [Google Scholar] [CrossRef]
- Nevo, Y.; Nelson, N. The NRAMP family of metal-ion transporters. BBA-Mol. Cell Res. 2006, 1763, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T.; et al. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef]
- Wu, D.; Yamaji, N.; Yamane, M.; Kashino-Fujii, M.; Sato, K.; Ma, J. The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol. 2016, 172, 1899–1910. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Bashir, K.; Senoura, T.; Sugimoto, K.; Ono, K.; Suzui, N.; Kawachi, N.; Ishii, S.; et al. From laboratory to field: OsNRAMP5-knockdown rice is a promising candidate for Cd phytoremediation in paddy fields. PLoS ONE 2014, 9, e98816. [Google Scholar] [CrossRef]
- Yamaji, N.; Sasaki, A.; Xia, J.X.; Yokosho, K.; Ma, J.F. A node-based switch for preferential distribution of manganese in rice. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [Green Version]
- Thomine, S.; Lelievre, F.; Debarbieux, E.; Schroeder, J.I.; Barbier-Brygoo, H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 2003, 34, 685–695. [Google Scholar] [CrossRef]
- Thomine, S.; Wang, R.; Ward, J.M.; Crawford, N.M.; Schroeder, J.I. Cadmium and iron transport by members of a plant transporter gene family in Arabidopsis with homology to NRAMP genes. Proc. Natl. Acad. Sci. USA 2000, 97, 4991–4996. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, X.; Peijnenburg, W.J.G.M.; Zhao, J.; Chen, X.; Yu, J.; Wu, H. Pathways of cadmium fluxes in the root of the halophyte Suaeda salsa. Ecotoxicol. Environ. Saf. 2012, 75, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFalco, T.A.; Marshall, C.B.; Munro, K.; Kang, H.G.; Moeder, W.; Ikura, M.; Snedden, W.A.; Yoshioka, K. Multiple calmodulin-binding sites positively and negatively regulate Arabidopsis CYCLIC NUCLEOTIDE-GATED CHANNEL12. Plant Cell 2016, 28, 1738–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, S.K.; Sharma, M.; Pandey, G.K. Role of cyclic nucleotide gated channels in stress management in plants. Curr. Genom. 2016, 17, 315–329. [Google Scholar] [CrossRef]
- Lam, H.M.; Chiu, J.; Hsieh, M.H.; Meisel, L.; Oliveira, I.C.; Shin, M.; Coruzzi, G. Glutamate-receptor genes in plants. Nature 1998, 396, 125–126. [Google Scholar] [CrossRef]
- Davenport, R. Glutamate receptors in plants. Ann. Bot. 2002, 90, 549–557. [Google Scholar] [CrossRef]
- Aouini, A.; Matsukura, C.; Ezura, H.; Asamizu, E. Characterisation of 13 glutamate receptor-like genes encoded in the tomato genome by structure, phylogeny and expression profiles. Gene 2012, 493, 36–43. [Google Scholar] [CrossRef]
- Ni, J.; Yu, Z.; Du, G.; Zhang, Y.; Taylor, J.L.; Shen, C.; Xu, J.; Liu, X.; Wang, Y.; Wu, Y. Heterologous expression and functional analysis of rice GLUTAMATE RECEPTOR-LIKE family indicates its role in glutamate triggered calcium flux in rice roots. Rice 2016, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Montanini, B.; Blaudez, D.; Jeandroz, S.; Sanders, D.; Chalot, M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: Improved signature and prediction of substrate specificity. BMC Genom. 2007, 8, 107. [Google Scholar] [CrossRef] [Green Version]
- Kawachi, M.; Kobae, Y.; Mimura, T.; Maeshima, M. Deletion of ahistidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008, 283, 8374–8383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbrosses-Fonrouge, A.G.; Voigt, K.; Schroder, A.; Arrivault, S.; Thomine, S.; Kramer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef] [PubMed]
- Kawachi, M.; Kobae, Y.; Kogawa, S.; Mimura, T.; Kramer, U.; Maeshima, M. Amino acid screening based on structural modeling identifies critical residues for the function, ion selectivity and structure of Arabidopsis MTP1. FEBS J. 2012, 279, 2339–2356. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Papierniak, A.; Kosieradzka, A.; Posyniak, E.; Maciaszczyk-Dziubinska, E.; Biskup, R.; Garbiec, A.; Marchewka, T. Cucumber metal tolerance protein CsMTP9 is a plasma membrane H+-coupled antiporter involved in the Mn2+ and Cd2+ efflux from root cells. Plant J. 2015, 84, 1045–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, D.; Sasaki, A.; Yamaji, N.; Miyaji, T.; Fujii, Y.; Takemoto, Y.; Moriyama, S.; Che, J.; Moriyama, Y.; Iwasaki, K.; et al. A polarly localized transporter for efficient manganese uptake in rice. Nat. Plants 2015, 1, 1–8. [Google Scholar] [CrossRef]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T. Mn tolerance in rice is mediated by MTP8.1 a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Gustin, J.L.; Lahner, B.; Persans, M.W.; Baek, D.; Yun, D.J.; Salt, D.E. The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyce cerevisiae. Plant J. 2004, 39, 237–251. [Google Scholar] [CrossRef]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef] [Green Version]
- Lekeux, G.; Crowet, J.; Nouet, C.; Joris, M.; Jadoul, A.; Bosman, B.; Carnol, M.; Motte, P.; Lins, L.; Galleni, M.; et al. Homology modeling and in vivo functional characterization of the zinc permeation pathway in a heavy metal P-type ATPase. J. Exp. Bot. 2019, 70, 329–341. [Google Scholar] [CrossRef]
- Uraguchi, S.; Kamiya, T.; Clemens, S.; Fujiwara, T. Charaterization of OsLCT1, a cadmium transporter from Indica rice (Oryza sativa). Physiol. Plant. 2014, 151, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Park, J.; Mendoza-Cozatl, D.G.; Suter-Grotemeyer, M.; Shim, D.; Hortensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, P.; Zanella, L.; De, P.A.; Di, L.D.; Cecchetti, V.; Falasca, G. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becher, M.; Talke, I.N.; Krall, L.; Kramer, U. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 2004, 37, 251–268. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [Green Version]
- Chao, D.Y.; Silva, A.; Baxter, I.; Huang, Y.S.; Nordborg, M.; Danku, J. Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1002923. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.J.; Xu, W. Heavy metal ATPase3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Korenkov, V.; Hirschi, K.; Crutchfield, J.D.; Wagner, G.J. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 2007, 226, 1379–1387. [Google Scholar] [CrossRef]
- Korenkov, V.; King, B.; Hirschi, K.; Wagner, G.J. Root-selective expression of AtCAX4 and AtCAX2 results in reduced lamina cadmium in field-grown Nicotiana tabacum L. Plant Biotechnol. J. 2009, 7, 219–226. [Google Scholar] [CrossRef]
- Migocka, M.; Kosieradzka, A.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Posyniak, E.; Garbiec, A.; Filleur, S. Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 2014, 66, 1001–1015. [Google Scholar] [CrossRef] [Green Version]
- Migocka, M.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Pozdzik, P.; Posyniak, E.; Garbiec, A.; Filleur, S. Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5367–5384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podar, D.; Scherer, J.; Noordally, Z.; Herzyk, P.; Nies, D.; Sanders, D. Metal selectivity determinants in a family of transition metal transporters. J. Biol. Chem. 2012, 287, 3185–3196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 transporter is involved in root to shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Ma, J. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopittke, P.M.; DeJonge, M.D.; Wang, P.; Mckenna, B.A.; Lombi, E.; Paterson, D.J. Laterally resolved speciation of arsenic in roots of wheat and rice using fluorescence-XANES imaging. New Phytol. 2013, 201, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Meyer, S.; De Angeli, A.; Nagy, R. Vacuolar transporters in their physiological context. Annu. Rev. Plant. Biol. 2012, 63, 183–213. [Google Scholar] [CrossRef]
- Mathys, W. The role of malate, oxalate, and mustard oil glucosides in the evolution of zinc resistance in herbage plants. Physiol. Plant. 1997, 40, 130–136. [Google Scholar] [CrossRef]
- Ortiz, D.F.; Kreppel, L.; Speiser, D.M.; Scheel, G.; McDonald, G.; Ow, D.W. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette type vacuolar membrane transporter. EMBO J. 1992, 11, 3491–3499. [Google Scholar] [CrossRef]
- Ortiz, D.F.; Ruscitti, T.; McCue, K.F.; Ow, D.W. Transport of metal- binding peptides by HMT1, a fission Yeast ABC-type vacuolar membrane protein. J. Biol. Chem. 1995, 270, 4721–4728. [Google Scholar] [CrossRef] [Green Version]
- Kuriakose, S.V.; Prasad, M.N.V. Cadmium stress affects seed germination and seedling growth in Sorghumbicolor (L.) Moench by changing the activities of hydrolyzing enzymes. Plant Growth Reg. 2008, 54, 143–156. [Google Scholar] [CrossRef]
- Sooksa-Nguan, T.; Yakubov, B.; Kozlovskyy, V.; Barkume, C.M.; Howe, K.J.; Thannhauser, T.W. Drosophila ABCtransporter, DmHMT-1, confers tolerance to cadmium. DmHMT-1 and its yeast homolog, SpHMT-1, are not essential for vacuolar phytochelatin sequestration. J. Biol. Chem. 2009, 284, 354–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vatamaniuk, O.K.; Bucher, E.A.; Sundaram, M.V.; Rea, P.A. CeHMT-1, a putative phytochelatin transporter, is required for cadmium tolerance in Caenorhab ditiselegans. J. Biol. Chem. 2005, 280, 23684–23690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.P.; Li, Z.S.; Drozdowicz, Y.M.; Hortensteiner, S.; Martinoia, E.; Rea, P.A. AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with AtMRP1. Plant Cell 1998, 10, 267–282. [Google Scholar] [PubMed] [Green Version]
- Park, J.; Song, W.Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Song, W.Y.; Mendoza-Cozatl, D.; Lee, Y.; Schroeder, J.; Ahn, S.N.; Lee, H.S. Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 2014, 37, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T. OsHMA3, a P-1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, M.; Shohag, M.J.; Tian, S.; Song, H.; Feng, Y. Enhanced expression of SaHMA3 plays critical roles in Cd hyperaccumulation and hypertolerance in Cd hyperaccumulator Sedum alfredii Hance. Planta 2016, 243, 577–589. [Google Scholar] [CrossRef]
- Oomen, R.J.; Wu, J.; Lelievre, F.; Blanchet, S.; Richaud, P.; Barbier-Brygoo, H. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009, 181, 637–650. [Google Scholar] [CrossRef]
- Baliardini, C.; Meyer, C.L.; Salis, P.; Saumitou-Laprade, P.; Verbruggen, N. CATION EXCHANGER1 cosegregates with cadmium tolerance in the metal hyperaccumulator Arabidopsis halleri and plays a role in limiting oxidative stress in Arabidopsis spp. Plant Physiol. 2015, 169, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Lanquar, V.; Lelièvre, F.; Bolte, S.; Hamès, C.; Alcon, C.; Neumann, D. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005, 24, 4041–4051. [Google Scholar] [CrossRef]
- Boominathan, R.; Doran, P.M. Organic acid complexation, heavy metal distribution and the effect of ATPase inhibition in hairy roots of hyperaccumulator plant species. J. Biotechnol. 2003, 101, 131–146. [Google Scholar] [CrossRef]
- Sun, R.L.; Zhou, Q.X.; Jin, C.X. Cadmium accumulation in relation to organic acids in leaves of Solanum nigrum L. as a newly found cadmium hyperaccumulator. Plant Soil 2006, 285, 125–134. [Google Scholar] [CrossRef]
- Ehsan, S.; Ali, S.; Noureen, S.; Mahmood, K.; Farid, M.; Ishaque, W.; Shakoor, M.B.; Rizwan, M. Citric acid assisted phytoremediation of cadmium by Brassica napus L. Ecotoxicol. Environ. Saf. 2014, 106, 164–172. [Google Scholar] [CrossRef]
- Yang, H.; Yang, Z.M.; Zhou, L.X.; Wong, J.W. Ability of Agrogyron elongatum to accumulate the single metal of cadmium, copper, nickel and lead and root exudation of organic acids. J. Environ. Sci. 2001, 13, 368–375. [Google Scholar]
- Price, M.B.; Kong, D.; Okumoto, S. Inter-subunit interactions between Glutamate-Like Receptors in Arabidopsis. Plant Signal. Behav. 2013, 12, e27034. [Google Scholar] [CrossRef] [Green Version]

| Leafy Vegetable Species | Range of Cd in Shoot (mg∙kg−1) | Mean or Range of Cd in Soil (mg∙kg−1) | Reference |
|---|---|---|---|
| Lactuca sativa L. | 5.8 to 9.1 | 0.12 to 0.31 | Baldantoni et al. [32] |
| Cichorium endivia L. | 0.61 to 3.80 | 0.13 to 0.51 | Baldantoni et al. [32] |
| Brassica pekinensis L. | 1.05 to 3.51 | 2.42 | Wang et al. [33] |
| Transporter | Localization | Function/Substrate | Reference |
|---|---|---|---|
| OsNRAMP1 | Roots and shoots (PM) | Influx of Cd, Al | [42] |
| OsZIP8 | Root (PM) | Influx of Zn, Fe | [52] |
| OsHMA3 | Roots (tonoplast) | Cd sequestration in root vacuoles | [52] |
| SnYSL3 | Vascular tissues and epidermal cells of the roots and stems (PM) | Transport of nicotianamine complexes containing Fe(II), Cu, Zn, and Cd | [41] |
| OsYSL2 | Vascular bundles, Roots (PM) | Influx of nicotianamine complexes containing Fe(II), Mn, Ni, and Cd | [77] |
| OsIRT1 | Roots (PM) | Uptake of Fe, Zn, Mn, and Cd | [78] |
| OsIRT2 | Roots (PM) | Uptake of Fe, Zn, Mn, and Cd | [78] |
| OsNRAMP5 | Roots (PM) | Uptake of Mn and Cd | [82] |
| HvNRAMP5 | Roots (PM) | Uptake of Mn and Cd | [83] |
| OsNRAMP3 | Vascular bundles, roots, leaves (tonoplast) | Uptake of Mn | [85] |
| AtNRAMP3 | Vascular bundles, roots, leaves (tonoplast) | Efflux of Fe and Cd | [87,88] |
| AtNRAMP4 | Vascular bundles, roots, leaves (tonoplast) | Efflux of Fe and Cd | [87,88] |
| AtNRAMP6 | Roots, young leaves (PM) | Influx of Mn, | [40] |
| AtCNGCs | Roots (PM) | Transporter for multiple cations | [89,90,91,92] |
| AtGLRs | Roots (PM) | Transporter for multiple cations | [89,90,93,94,95,96] |
| AtMTP1 | Roots and leaves (tonoplast) | Transporter for Zn and Cd | [97,98,99,100] |
| CsMTP9 | Roots endodermal cells (PM) | Efflux of Mn and Cd | [101] |
| OsMTP9 | Roots (PM) | Efflux of Mn | [102] |
| OsMTP8.1 | Roots (tonoplast) | Sequestration of Mn into vacuoles | [103] |
| TgMTP1 | Roots and leaves (tonoplast) | Transporter for Zn and Cd | [104] |
| OsHMA5 | Roots, vascular bundles (tonoplast) | Loading of Cu in xylem | [105] |
| AtHMA2 | Roots, vascular tissue (PM) | Delivery of Zn and Cd to xylem | [106,107] |
| AtHMA4 | Roots, vascular tissue (PM) | Delivery of Zn and Cd to xylem | [106,107] |
| OsLCT1 | Leaves, nodes, phloem parenchyma (PM) | Efflux of Cd, Ca, Mg, and Mn | [74,108] |
| AtABCC1 | Roots and shoots (tonoplast) | Uptake of PCs | [109] |
| AtABCC2 | Roots and shoots (tonoplast) | Uptake of PCs | [109] |
| AtABCC3 | Roots and shoots (tonoplast) | Uptake of PCs | [110] |
| AhHMA3 | Roots, shoots (tonoplast) | Sequestration of Zn into vacuoles | [111] |
| AtHMA3 | Vascular tissues (tonoplast) | Transport of Zn, Co, Pb, and Cd | [112,113] |
| SpHMA3 | Roots, shoots (tonoplast) | Sequestration of Cd into vacuoles | [114] |
| AtCAX2 | Roots (tonoplast) | Vacuolar Cd, Zn, and Mn transport | [115,116] |
| AtCAX4 | Roots (tonoplast) | Vacuolar Cd, Zn, and Mn transport | [115,116] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Duan, S.; Wu, Q.; Yu, M.; Shabala, S. Reducing Cadmium Accumulation in Plants: Structure–Function Relations and Tissue-Specific Operation of Transporters in the Spotlight. Plants 2020, 9, 223. https://doi.org/10.3390/plants9020223
Huang X, Duan S, Wu Q, Yu M, Shabala S. Reducing Cadmium Accumulation in Plants: Structure–Function Relations and Tissue-Specific Operation of Transporters in the Spotlight. Plants. 2020; 9(2):223. https://doi.org/10.3390/plants9020223
Chicago/Turabian StyleHuang, Xin, Songpo Duan, Qi Wu, Min Yu, and Sergey Shabala. 2020. "Reducing Cadmium Accumulation in Plants: Structure–Function Relations and Tissue-Specific Operation of Transporters in the Spotlight" Plants 9, no. 2: 223. https://doi.org/10.3390/plants9020223
APA StyleHuang, X., Duan, S., Wu, Q., Yu, M., & Shabala, S. (2020). Reducing Cadmium Accumulation in Plants: Structure–Function Relations and Tissue-Specific Operation of Transporters in the Spotlight. Plants, 9(2), 223. https://doi.org/10.3390/plants9020223





