SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family: TRANSCRIPTOME-Wide Identification, Phylogenetic Relationship, Expression Patterns and Network Interaction Analysis in Panax ginseng C. A. Meyer
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the Ginseng SPL Transcripts
2.2. GO (Gene Ontology) Annotation, Functional Categorization and Analyses
2.3. Multiple Sequence Alignments and Phylogenetic Analysis
2.4. Ka/Ks Ratio among SPL in Ginseng and Other Species
2.5. Analysis of Different PgSPL Groups for Conserved Motifs
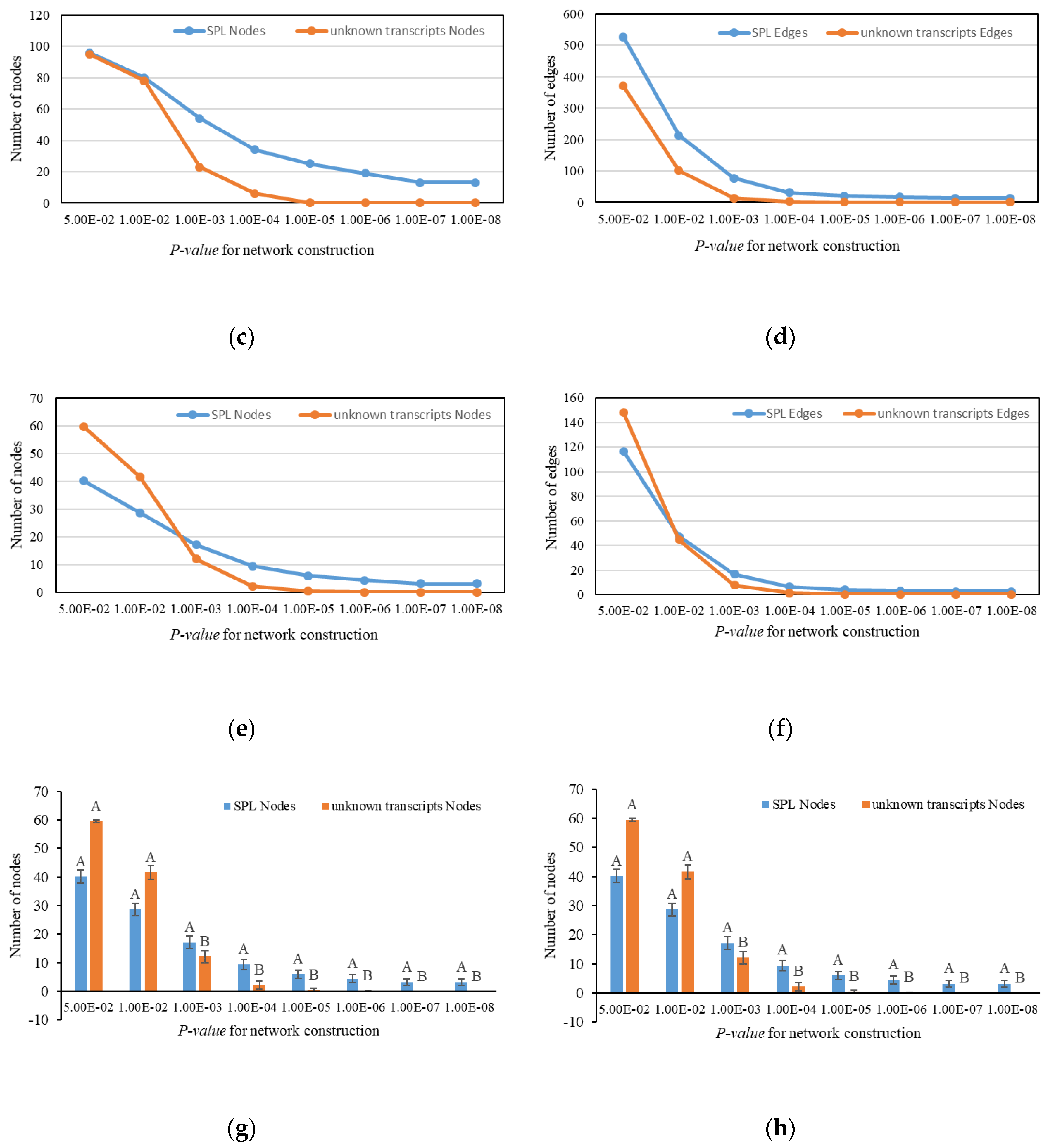
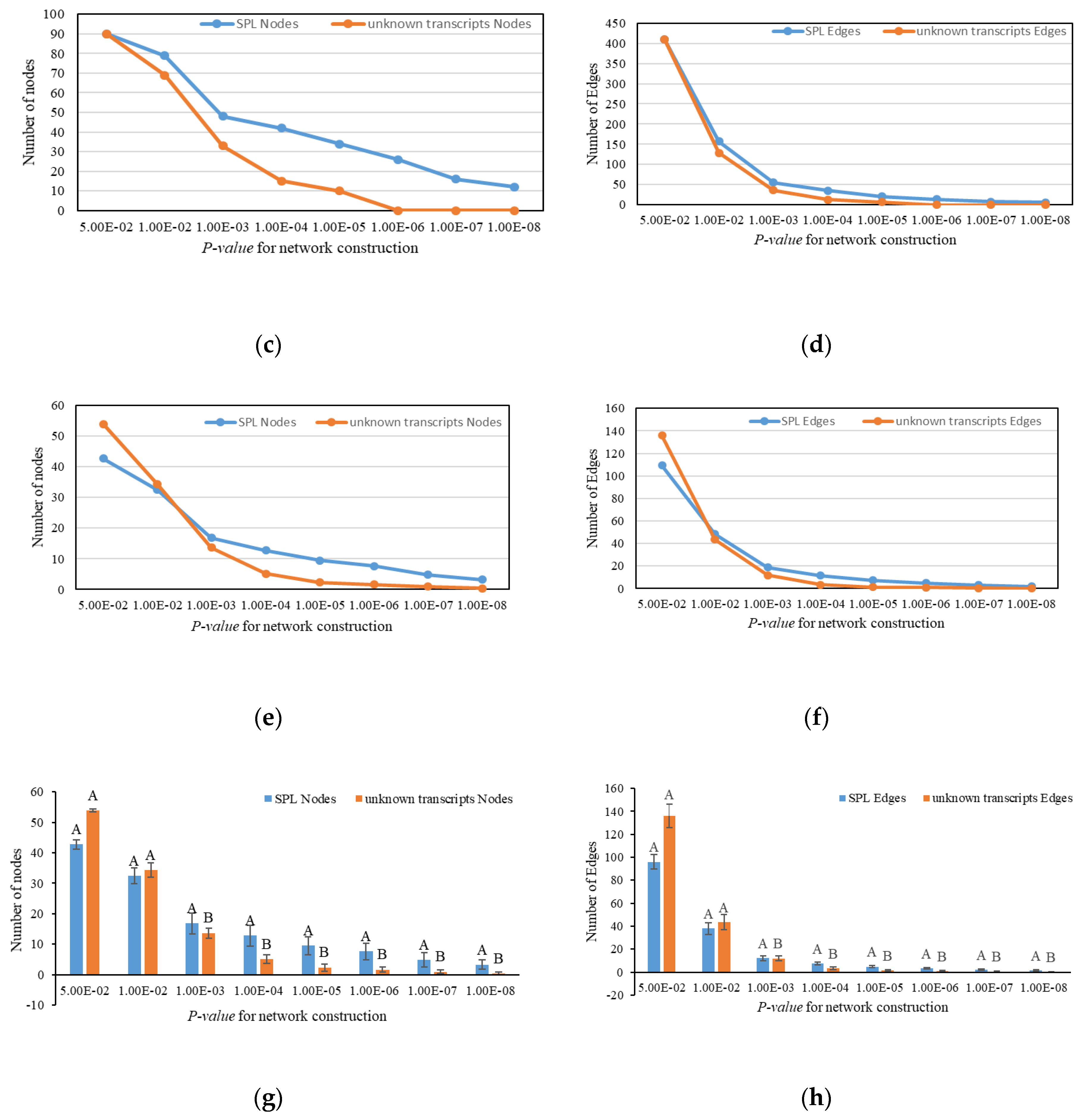
2.6. Expression and Interaction Characteristic of PgSPL Transcripts
3. Results
3.1. Identification of the Ginseng SPL Transcripts
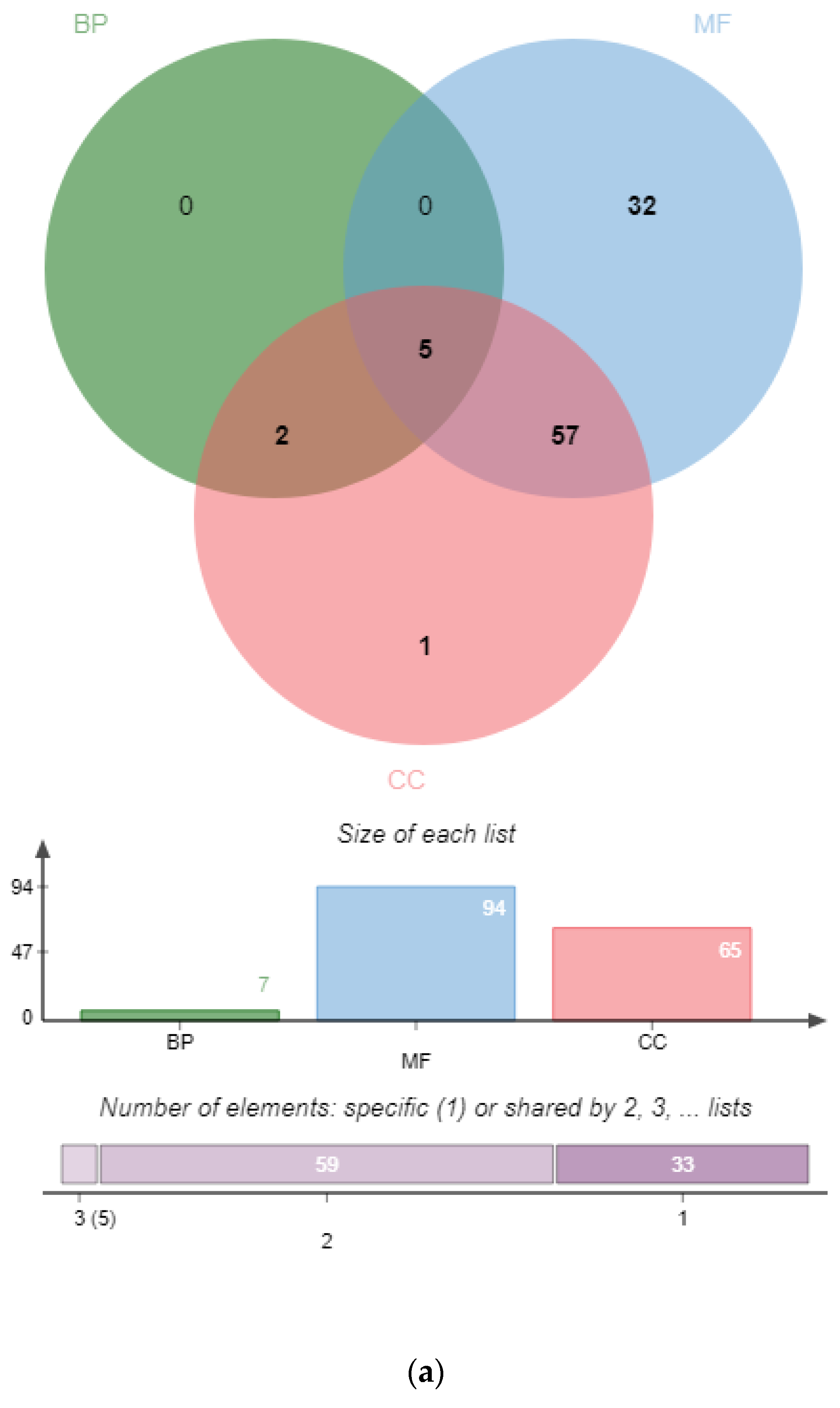
3.2. GO Annotation of PgSPL Transcripts
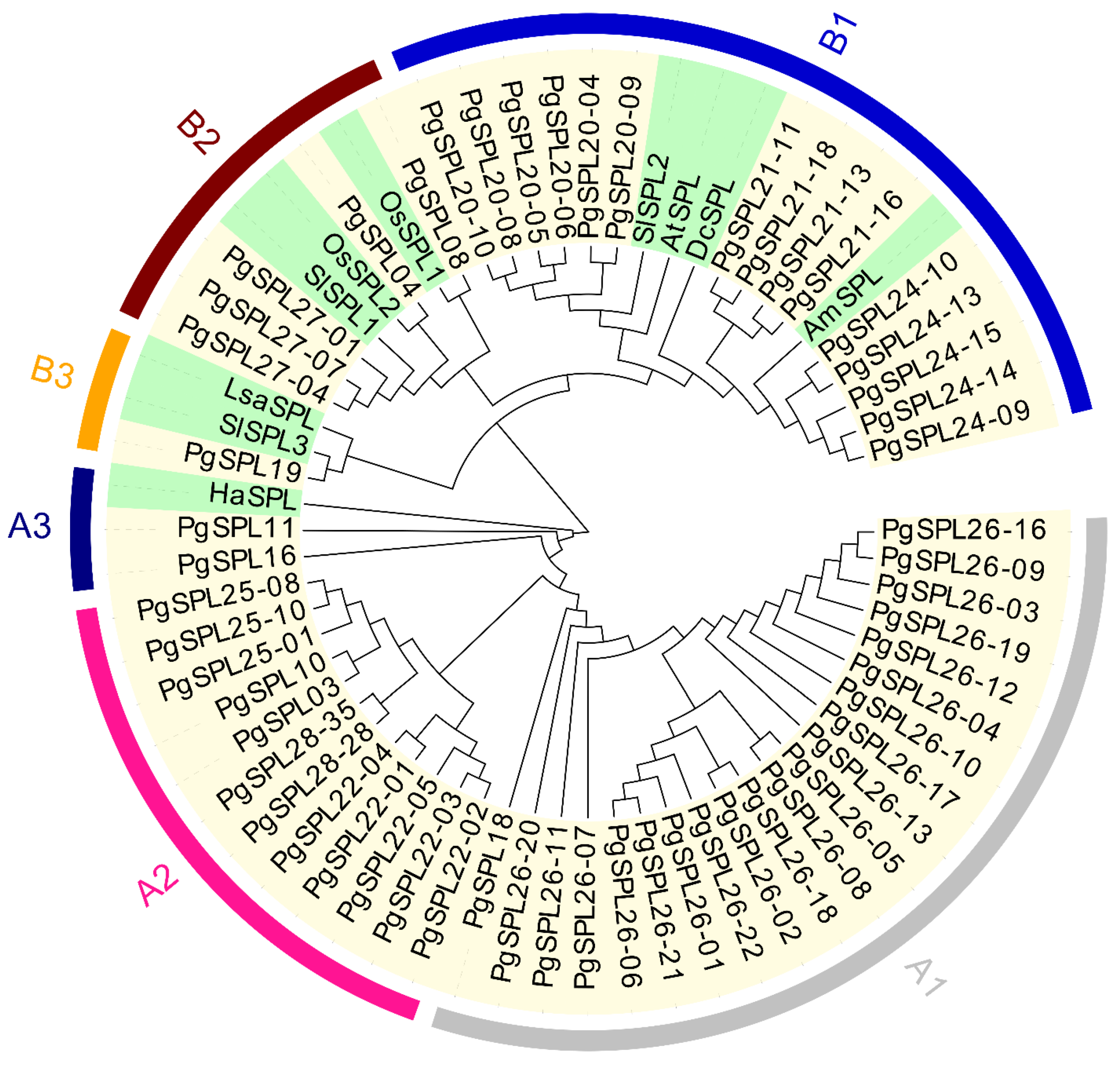
3.3. Phylogenetic Relationship and Classification for PgSPL Transcripts
3.4. Synonymous and Nonsynonymous Substitution Rates and Selective Pressure in PgSPL
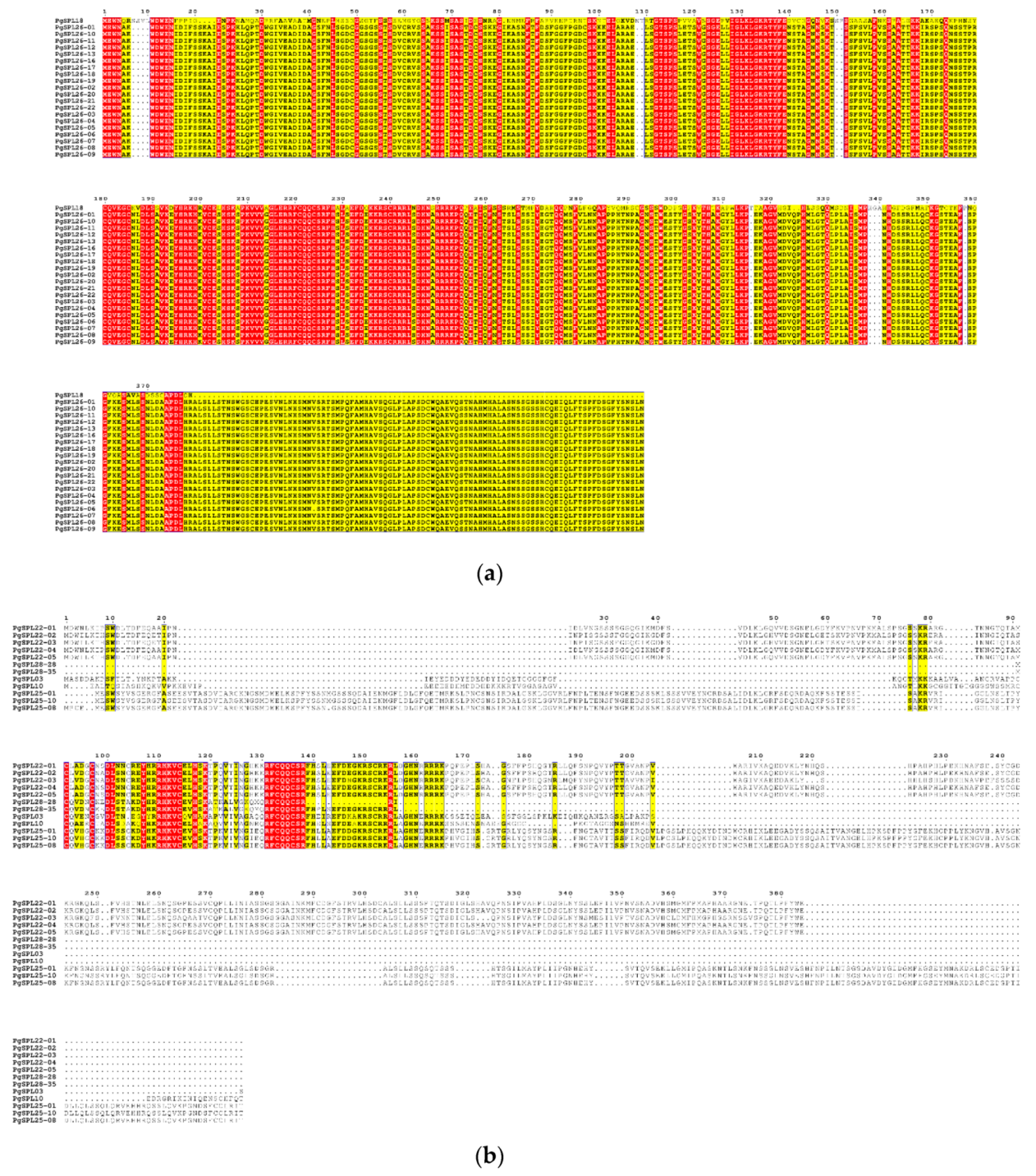
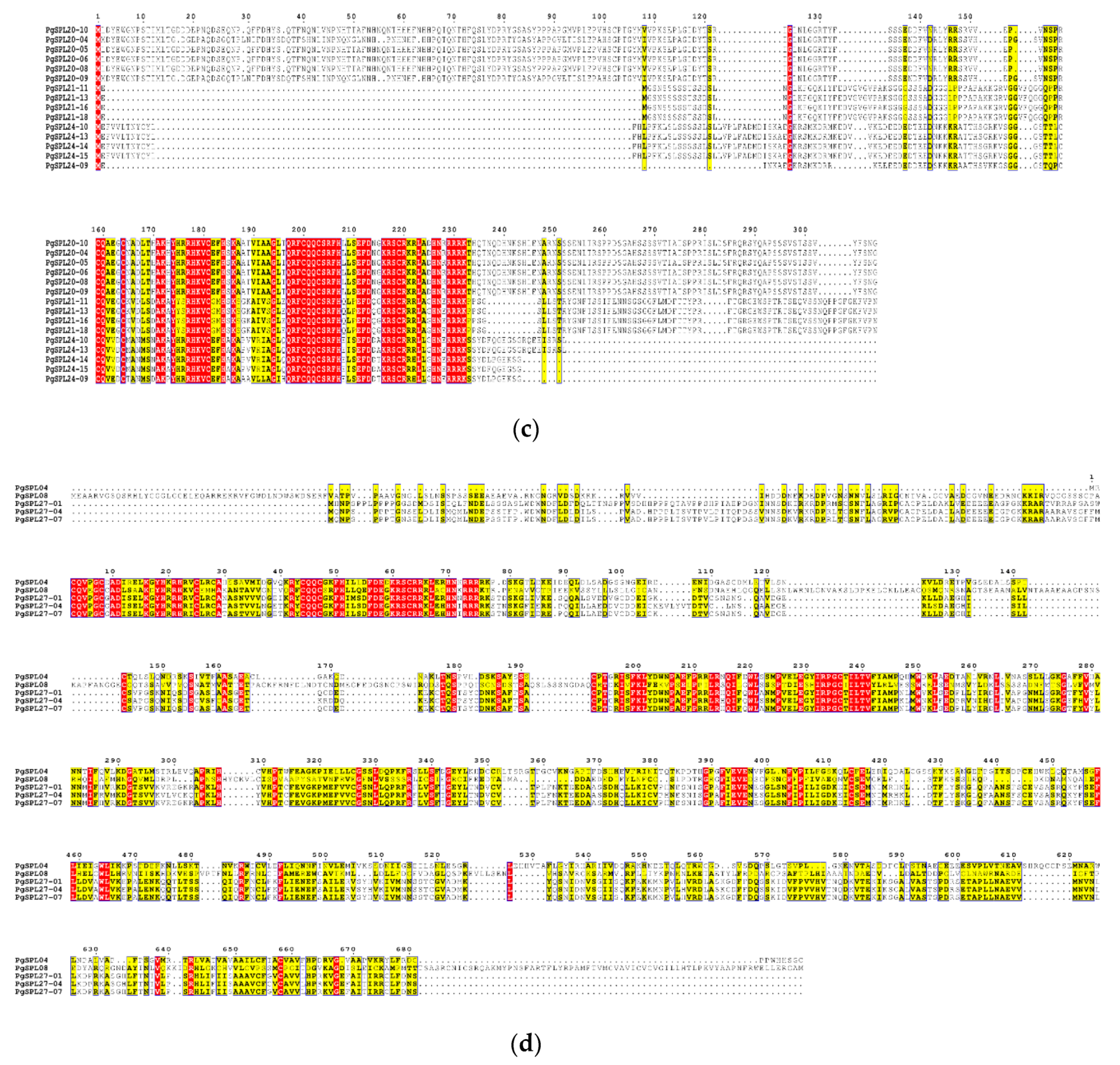
3.5. Analysis for Conserved Domains of PgSPL Transcripts
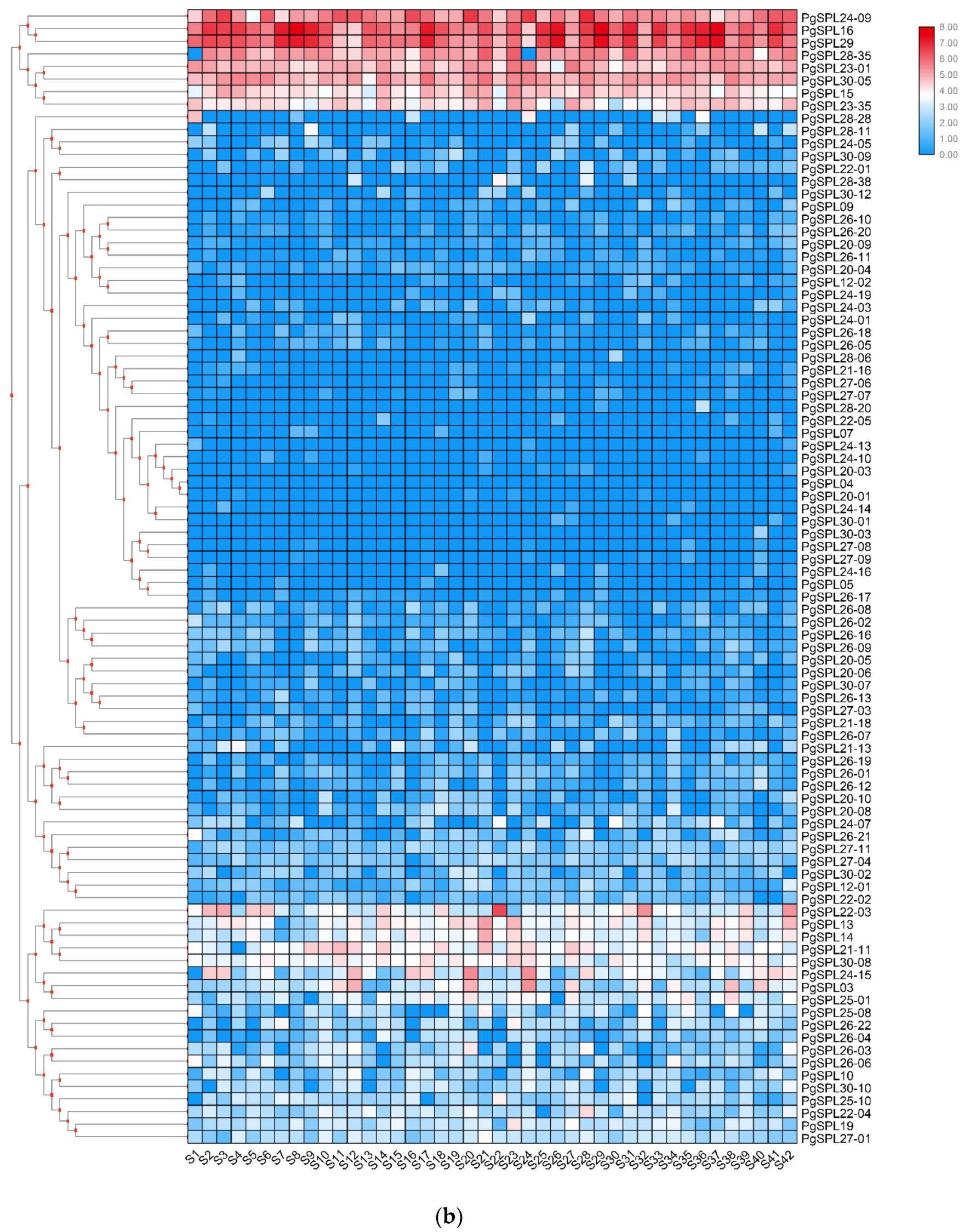
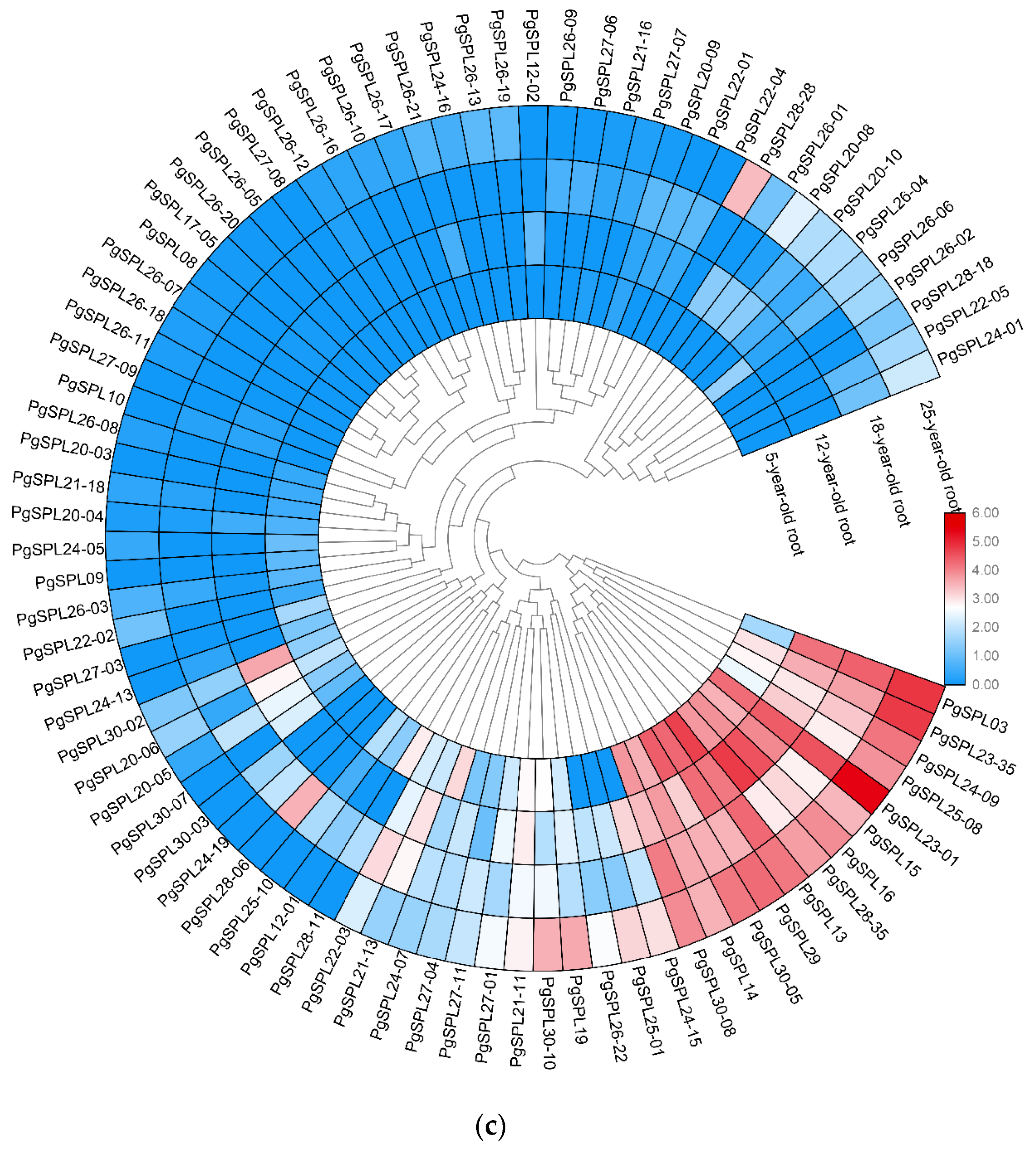
3.6. Temporal and Spatial Expression Characteristic of PgSPL Transcripts
3.7. Expression Characteristic of PgSPL Transcripts in 4-Year-Old Roots of Different Genotypes
4. Discussion
4.1. SPL Genes in Ginseng is a Biggish Gene Family
4.2. PgSPL Transcripts Have a Relatively Centralized Distribution of Functions and Tend to Serve for Close Purposes
4.3. Some PgSPL Transcripts with a Prominent Spatial Expression Pattern
4.4. Some PgSPL Transcripts with a Prominent Temporal Expression Pattern
4.5. Some PgSPL Transcripts with a Prominent Genotype Expression Pattern
4.6. PgSPL as Ginseng’s Age Molecular Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Additional Information
References
- Luscombe, N.M.; Austin, S.E.; Berman, H.M.; Thornton, J.M. An overview of the structures of protein–DNA complexes. Genome Biol. 2000, 1, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar]
- Riese, M.; Zobell, O.; Saedler, H.; Huijser, P. SBP-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss Physcomitrella patens. Planta 2008, 227, 505–515. [Google Scholar] [CrossRef]
- Cardon, G.H.; Höhmann, S.; Nettesheim, K.; Saedler, H.; Huijser, P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997, 12, 367–377. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.J.; Lu, Q.; Wilson, I.W.; Qiu, D.Y. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujube. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL transcripts and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Hohmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-Box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Bikenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, J.W. Recent Progress in miR156-mediated aging pathway in plants. Chin. Sci. Bull. 2014, 59, 1398–1404. (In Chinese) [Google Scholar]
- Ahuja, A.; Kim, J.H.; Kim, J.-H.; Yi, Y.-S.; Cho, J.Y. Functional role of ginseng-derived compounds in cancer. J. Ginseng Res. 2018, 42, 248–254. [Google Scholar] [CrossRef]
- Irfan, M.; Kim, M.; Rhee, M.H. Anti-platelet role of Korean ginseng and ginsenosides in cardiovascular diseases. J. Ginseng Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-I.; Park, K.S.; Cho, I.-H. Panax ginseng: A candidate herbal medicine for autoimmune disease. J. Ginseng Res. 2019, 43, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.-S. Ameliorative effects of ginseng and ginsenosides on rheumatic diseases. J. Ginseng Res. 2019, 43, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.S.; Auyeung, K.K.; Yip, K.M.; Ye, R.; Zhao, Z.Z.; Mao, Q.; Xu, J.; Chen, H.B.; Li, S.L. Stronger anti-obesity effect of white ginseng over red ginseng and the potential mechanisms involving chemically structural/compositional specificity to gut microbiota. Phytomedicine 2018, 19, 152761. [Google Scholar] [CrossRef]
- Kim, J.; Han, S.Y.; Min, H. Ginseng for an eye: Effects of ginseng on ocular diseases. J. Ginseng Res. 2018. [Google Scholar] [CrossRef]
- Nishijo, H.; Uwano, T.; Zhong, Y.-M.; Ono, T. Proof of the mysterious efficacy of ginseng: Basic and clinical trials: Effects of red ginseng on learning and memory deficits in an animal model of amnesia. J. Pharmacol. Sci. 2004, 95, 145–152. [Google Scholar] [CrossRef]
- Lee, M.J.; Kim, E.H.; Rhee, D.K. Effects of Panax ginseng on stress. J. Ginseng Res. 2008, 32, 8–14. [Google Scholar]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Javier, T.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Xu, B.; Yang, Z. PAMLX: A graphical user interface for PAML. Mol. Biol. Evol. 2013, 30, 2723–2724. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; He, Y.H.; Xia, R. TBtools, a Toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Freeman, T.C.; Goldovsky, L.; Brosch, M.; van Dongen, S.; Mazière, P.; Grocock, R.J.; Freilich, S.; Thornton, J.; Enright, A.J. Construction, visualisation and clustering of transcription networks from microarray expression data. PLoS Comput. Biol. 2007, 3, e206. [Google Scholar] [CrossRef]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-Regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Chuck, G.; Cigan, A.M.; Saeteurn, K.; Hake, S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 2007, 39, 544–549. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. miRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Somoza, I.R.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef]







| Gene | Transcript | Seq. Length (bp) | ORF Length (aa) | MW (kDa) | pI |
|---|---|---|---|---|---|
| PgSPL01 | PgSPL01 | 231 | 65 | 7.22 | 8.79 |
| PgSPL02 | PgSPL02 | 529 | 88 | 10.08 | 4.81 |
| PgSPL03 | PgSPL03 | 988 | 182 | 20.32 | 8.74 |
| PgSPL04 | PgSPL04 | 2342 | 692 | 77.39 | 6.76 |
| PgSPL05 | PgSPL05 | 280 | 76 | 9.28 | 10.73 |
| PgSPL06 | PgSPL06 | 254 | 76 | 8.83 | 9.84 |
| PgSPL07 | PgSPL07 | 216 | 58 | 6.53 | 8.88 |
| PgSPL08 | PgSPL08 | 3216 | 961 | 105.47 | 5.67 |
| PgSPL09 | PgSPL09 | 264 | 81 | 8.81 | 9.64 |
| PgSPL10 | PgSPL10 | 1037 | 199 | 21.92 | 8.83 |
| PgSPL11 | PgSPL11 | 3582 | 966 | 106.26 | 6.82 |
| PgSPL12 | PgSPL12-01 | 511 | 63 | 6.56 | 4.58 |
| PgSPL12-02 | 523 | 66 | 6.80 | 4.58 | |
| PgSPL13 | PgSPL13 | 832 | 74 | 8.56 | 11.40 |
| PgSPL14 | PgSPL14 | 1097 | 210 | 22.96 | 9.03 |
| PgSPL15 | PgSPL15 | 439 | 67 | 7.28 | 4.33 |
| PgSPL16 | PgSPL16 | 930 | 198 | 22.16 | 8.91 |
| PgSPL17 | PgSPL17-01 | 4180 | 264 | 29.47 | 6.25 |
| PgSPL17-04 | 4149 | 307 | 35.33 | 6.19 | |
| PgSPL17-05 | 3882 | 264 | 29.47 | 6.25 | |
| PgSPL18 | PgSPL18 | 1146 | 378 | 41.28 | 9.52 |
| PgSPL19 | PgSPL19 | 684 | 182 | 20.82 | 9.77 |
| PgSPL20 | PgSPL20-01 | 982 | 65 | 7.47 | 5.68 |
| PgSPL20-10 | 994 | 307 | 34.87 | 8.66 | |
| PgSPL20-03 | 1779 | 65 | 7.47 | 5.68 | |
| PgSPL20-04 | 1697 | 307 | 34.33 | 8.72 | |
| PgSPL20-05 | 1685 | 307 | 34.87 | 8.66 | |
| PgSPL20-06 | 1786 | 307 | 34.87 | 8.66 | |
| PgSPL20-08 | 1791 | 307 | 34.87 | 8.66 | |
| PgSPL20-09 | 1798 | 307 | 34.33 | 8.72 | |
| PgSPL21 | PgSPL21-11 | 2176 | 213 | 22.94 | 9.78 |
| PgSPL21-13 | 2164 | 213 | 22.94 | 9.78 | |
| PgSPL21-16 | 2206 | 213 | 22.94 | 9.78 | |
| PgSPL21-18 | 2218 | 213 | 22.94 | 9.78 | |
| PgSPL22 | PgSPL22-01 | 1827 | 384 | 42.24 | 8.26 |
| PgSPL22-02 | 1751 | 384 | 42.25 | 8.01 | |
| PgSPL22-03 | 1691 | 380 | 41.86 | 7.61 | |
| PgSPL22-04 | 1827 | 384 | 42.24 | 8.26 | |
| PgSPL22-05 | 1735 | 384 | 42.26 | 8.26 | |
| PgSPL23 | PgSPL23-01 | 1046 | 247 | 27.33 | 7.64 |
| PgSPL23-35 | 454 | 70 | 7.95 | 9.50 | |
| PgSPL24 | PgSPL24-01 | 531 | 68 | 7.79 | 6.79 |
| PgSPL24-10 | 1312 | 186 | 21.35 | 8.77 | |
| PgSPL24-11 | 1968 | 126 | 14.21 | 6.95 | |
| PgSPL24-13 | 1211 | 186 | 21.35 | 8.77 | |
| PgSPL24-14 | 776 | 177 | 20.27 | 8.76 | |
| PgSPL24-15 | 1783 | 177 | 20.25 | 8.77 | |
| PgSPL24-16 | 680 | 95 | 11.58 | 9.35 | |
| PgSPL24-19 | 1026 | 68 | 7.79 | 6.79 | |
| PgSPL24-03 | 1155 | 61 | 6.87 | 9.94 | |
| PgSPL24-05 | 1127 | 95 | 11.58 | 9.35 | |
| PgSPL24-07 | 1187 | 58 | 6.68 | 10.23 | |
| PgSPL24-09 | 430 | 136 | 15.59 | 8.68 | |
| PgSPL25 | PgSPL25-01 | 2764 | 533 | 58.86 | 8.29 |
| PgSPL25-10 | 2408 | 533 | 58.86 | 8.29 | |
| PgSPL25-08 | 2606 | 537 | 59.32 | 8.10 | |
| PgSPL26 | PgSPL26-01 | 2025 | 469 | 50.92 | 8.14 |
| PgSPL26-10 | 2377 | 469 | 50.90 | 8.14 | |
| PgSPL26-11 | 1844 | 469 | 50.90 | 8.14 | |
| PgSPL26-12 | 1932 | 469 | 50.90 | 8.14 | |
| PgSPL26-13 | 2484 | 469 | 50.90 | 8.14 | |
| PgSPL26-16 | 2044 | 469 | 50.90 | 8.14 | |
| PgSPL26-17 | 2391 | 469 | 50.90 | 8.14 | |
| PgSPL26-18 | 2591 | 469 | 50.92 | 8.14 | |
| PgSPL26-19 | 1860 | 469 | 50.90 | 8.14 | |
| PgSPL26-02 | 2288 | 469 | 50.92 | 8.14 | |
| PgSPL26-20 | 2407 | 469 | 50.90 | 8.14 | |
| PgSPL26-21 | 1948 | 469 | 50.92 | 8.14 | |
| PgSPL26-22 | 2177 | 469 | 50.92 | 8.14 | |
| PgSPL26-03 | 1955 | 469 | 50.90 | 8.14 | |
| PgSPL26-04 | 2193 | 469 | 50.90 | 8.14 | |
| PgSPL26-05 | 2270 | 469 | 50.92 | 8.14 | |
| PgSPL26-06 | 2043 | 469 | 50.92 | 8.14 | |
| PgSPL26-07 | 2502 | 469 | 50.90 | 8.14 | |
| PgSPL26-08 | 1937 | 469 | 50.92 | 8.14 | |
| PgSPL26-09 | 2132 | 469 | 50.90 | 8.14 | |
| PgSPL27 | PgSPL27-01 | 3029 | 795 | 88.78 | 6.53 |
| PgSPL27-11 | 2821 | 446 | 50.05 | 7.56 | |
| PgSPL27-03 | 2879 | 447 | 50.07 | 7.57 | |
| PgSPL27-04 | 2886 | 791 | 88.44 | 6.61 | |
| PgSPL27-06 | 2864 | 446 | 50.05 | 7.56 | |
| PgSPL27-07 | 2828 | 786 | 87.97 | 6.32 | |
| PgSPL27-08 | 2806 | 446 | 50.05 | 7.56 | |
| PgSPL27-09 | 3051 | 446 | 50.05 | 7.56 | |
| PgSPL28 | PgSPL28-11 | 467 | 110 | 12.45 | 8.97 |
| PgSPL28-13 | 614 | 83 | 9.54 | 10.51 | |
| PgSPL28-18 | 509 | 247 | 27.25 | 5.74 | |
| PgSPL28-19 | 480 | 120 | 13.28 | 8.97 | |
| PgSPL28-20 | 1085 | 249 | 27.64 | 6.32 | |
| PgSPL28-27 | 680 | 120 | 13.28 | 8.97 | |
| PgSPL28-28 | 1092 | 50 | 5.90 | 9.40 | |
| PgSPL28-03 | 693 | 72 | 8.27 | 5.25 | |
| PgSPL28-30 | 509 | 111 | 12.48 | 8.97 | |
| PgSPL28-35 | 575 | 65 | 7.84 | 9.42 | |
| PgSPL28-38 | 538 | 111 | 12.48 | 8.97 | |
| PgSPL28-06 | 588 | 119 | 13.25 | 8.97 | |
| PgSPL29 | PgSPL29 | 3288 | 828 | 91.11 | 6.17 |
| PgSPL30 | PgSPL30-01 | 1106 | 157 | 17.03 | 5.68 |
| PgSPL30-10 | 878 | 113 | 12.89 | 8.47 | |
| PgSPL30-12 | 695 | 104 | 11.25 | 5.40 | |
| PgSPL30-02 | 1631 | 113 | 12.89 | 8.47 | |
| PgSPL30-03 | 1414 | 90 | 10.33 | 9.10 | |
| PgSPL30-05 | 1328 | 210 | 22.32 | 5.79 | |
| PgSPL30-07 | 1162 | 210 | 22.32 | 5.79 | |
| PgSPL30-08 | 998 | 157 | 17.03 | 5.68 | |
| PgSPL30-09 | 859 | 157 | 16.54 | 5.51 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Li, L.; Jiang, Y.; Wu, J.; Sun, H.; Zhao, M.; Jiang, Y.; Zhu, L.; Wang, Y.; Su, Y.; et al. SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family: TRANSCRIPTOME-Wide Identification, Phylogenetic Relationship, Expression Patterns and Network Interaction Analysis in Panax ginseng C. A. Meyer. Plants 2020, 9, 354. https://doi.org/10.3390/plants9030354
Li S, Li L, Jiang Y, Wu J, Sun H, Zhao M, Jiang Y, Zhu L, Wang Y, Su Y, et al. SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family: TRANSCRIPTOME-Wide Identification, Phylogenetic Relationship, Expression Patterns and Network Interaction Analysis in Panax ginseng C. A. Meyer. Plants. 2020; 9(3):354. https://doi.org/10.3390/plants9030354
Chicago/Turabian StyleLi, Shaokun, Li Li, Yang Jiang, Jun Wu, Honghua Sun, Mingzhu Zhao, Yue Jiang, Lei Zhu, Yanfang Wang, Yingjie Su, and et al. 2020. "SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family: TRANSCRIPTOME-Wide Identification, Phylogenetic Relationship, Expression Patterns and Network Interaction Analysis in Panax ginseng C. A. Meyer" Plants 9, no. 3: 354. https://doi.org/10.3390/plants9030354
APA StyleLi, S., Li, L., Jiang, Y., Wu, J., Sun, H., Zhao, M., Jiang, Y., Zhu, L., Wang, Y., Su, Y., Wang, K., Wang, Y., & Zhang, M. (2020). SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family: TRANSCRIPTOME-Wide Identification, Phylogenetic Relationship, Expression Patterns and Network Interaction Analysis in Panax ginseng C. A. Meyer. Plants, 9(3), 354. https://doi.org/10.3390/plants9030354






