AtXRN4 Affects the Turnover of Chosen miRNA*s in Arabidopsis
Abstract
:1. Introduction
2. Results
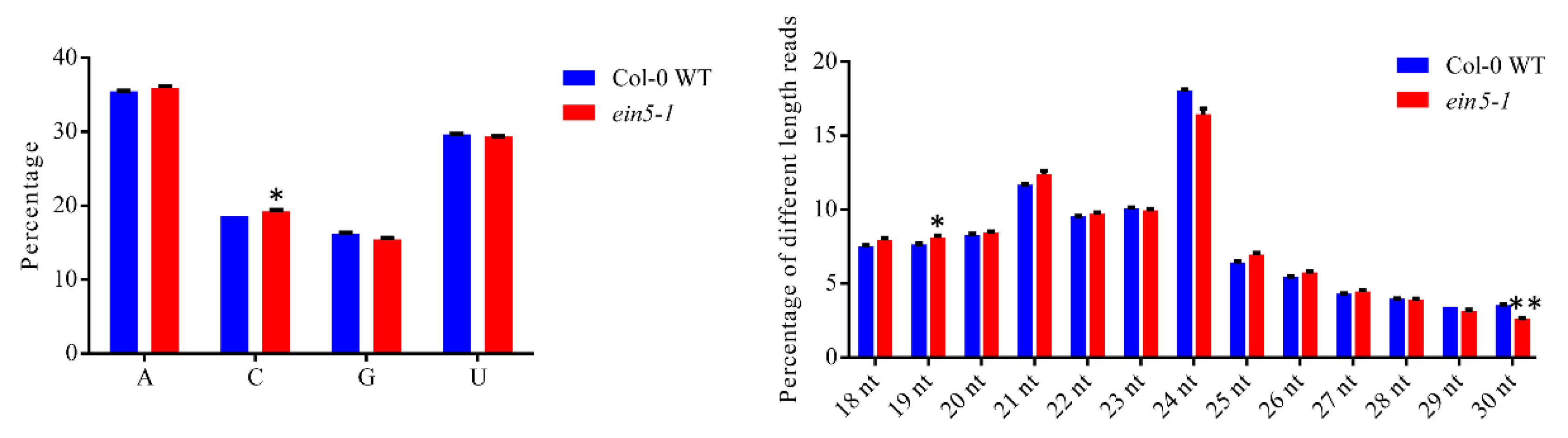
2.1. AtXRN4 Mutation Changes the sRNA Profiles
2.2. AtXRN4 Decreases the Accumulation of miRNA*s
2.3. AtXRN4 does not Alter theTranscription and Processing of miRNAsPrecursors
2.4. AtXRN2 and AtXRN3 do not Alter the Accumulations of Corresponding miRNA*s
2.5. AtXRN4 Interacts with AtAGO2 in P-Bodies
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and sRNASequencing
4.3. sRNA Sequencing Analysis
4.4. qRT-PCR
4.5. Northern Blot
4.6. Plasmid Construction
4.7. BiFC and Colocalization Assay
4.8. Co-IP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
References
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727. [Google Scholar] [CrossRef] [Green Version]
- Sanei, M.; Chen, X. Mechanisms of microRNA turnover. Curr. Opin. Plant Biol. 2015, 27, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Rüegger, S.; Großhans, H. MicroRNA turnover: When, how, and why. Trends Biochem. Sci. 2012, 37, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr. Biol. CB 2005, 15, 1501–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, G.; Chen, X.; Yu, B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. CB 2012, 22, 695–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, S.; Dou, Y.; Zhang, C.; Chen, X.; Yu, B.; Ren, G. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3’ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015, 11, e1005091. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Y.; Dou, Y.; Chen, L.; Wang, J.; Jiang, N.; Guo, C.; Yao, Q.; Wang, C.; Liu, L.; et al. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3’ to 5’ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E6659–E6667. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, V.; Chen, X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 2008, 321, 1490–1492. [Google Scholar] [CrossRef] [Green Version]
- Tops, B.B.; Tabara, H.; Sijen, T.; Simmer, F.; Mello, C.C.; Plasterk, R.H.; Ketting, R.F. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 2005, 33, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bail, S.; Swerdel, M.; Liu, H.; Jiao, X.; Goff, L.A.; Hart, R.P.; Kiledjian, M. Differential regulation of microRNA stability. RNA 2010, 16, 1032–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.F.; L’Etoile, N.D.; Ansel, K.M. Eri1: A conserved enzyme at the crossroads of multiple RNA-processing pathways. Trends Genet. TIG 2014, 30, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Fasler, M.; Büssing, I.; Großhans, H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell 2011, 20, 388–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Großhans, H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 2009, 461, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e4. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y. Activity and roles of Arabidopsis thaliana XRN family exoribonucleases in noncoding RNA pathways. J. Plant Res. 2017, 130, 25–31. [Google Scholar] [CrossRef]
- Kastenmayer, J.; Green, P. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 2000, 97, 13985–13990. [Google Scholar] [CrossRef] [Green Version]
- Roman, G.; Lubarsky, B.; Kieber, J.J.; Rothenberg, M.; Ecker, J.R. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 1995, 139, 1393–1409. [Google Scholar]
- Olmedo, G.; Guo, H.; Gregory, B.D.; Nourizadeh, S.D.; Aguilar-Henonin, L.; Li, H.; An, F.; Guzman, P.; Ecker, J.R. ETHYLENE-INSENSITIVE5 encodes a 5’→3’ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 2006, 103, 13286–13293. [Google Scholar] [CrossRef] [Green Version]
- Potuschak, T.; Vansiri, A.; Binder, B.M.; Lechner, E.; Vierstra, R.D.; Genschik, P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 2006, 18, 3047–3057. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souret, F.F.; Kastenmayer, J.P.; Green, P.J. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 2004, 15, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jones, C.I.; Newbury, S.F.; Green, P.J. XRN 5’→3’ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta 2013, 1829, 590–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazzani, S.; Lawrenson, T.; Woodward, C.; Headon, D.; Sablowski, R. A link between mRNA turnover and RNA interference in Arabidopsis. Science 2004, 306, 1046–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhu, Y.; Liu, X.; Hong, X.; Xu, Y.; Zhu, P.; Shen, Y.; Wu, H.; Ji, Y.; Wen, X.; et al. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 2015, 348, 120–123. [Google Scholar] [CrossRef]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Gregory, B.D.; O’Malley, R.C.; Lister, R.; Urich, M.A.; Tonti-Filippini, J.; Chen, H.; Millar, A.H.; Ecker, J.R. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 2008, 14, 854–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, F.; Guo, G.; Du, J.; Guo, W.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Xu, Z.; Zhao, W.; Liu, Q.; Li, Q.; Lu, L.; Liu, R.; Zhang, X.; Cui, F. Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus. BMC Plant Biol. 2018, 18, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef]
- Liu, C.; Axtell, M.J.; Fedoroff, N.V. The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 2012, 159, 748–758. [Google Scholar] [CrossRef] [Green Version]
- You, C.; He, W.; Hang, R.; Zhang, C.; Cao, X.; Guo, H.; Chen, X.; Cui, J.; Mo, B. FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun. 2019, 10, 4424. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. Cell Mol. Biol. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Meyer, S.; Temme, C.; Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Till, D.D.; Linz, B.; Seago, J.E.; Elgar, S.J.; Marujo, P.E.; de Lourdes Elias, M.; Arraiano, C.M.; McClellan, J.A.; McCarthy, J.E.; Newbury, S.F. Identification and developmental expression of a 5′-3′ exoribonuclease from Drosophila melanogaster. Mech. Dev. 1998, 79, 51–55. [Google Scholar] [CrossRef]
- West, S.; Gromak, N.; Proudfoot, N.J. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 2004, 432, 522. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5’-mononucleotides by a 5’leads to 3’mode of hydrolysis. J. Biol. Chem. 1980, 255, 3080–3085. [Google Scholar] [PubMed]
- Bashkirov, V.I.; Scherthan, H.; Solinger, J.A.; Buerstedde, J.-M.; Heyer, W.-D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997, 136, 761–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NEWBURY, S.; Woollard, A. The 5′-3′ exoribonuclease xrn-1 is essential for ventral epithelial enclosure during, C. elegans embryogenesis. RNA 2004, 10, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [Green Version]
- Bossé, G.D.; Rüegger, S.; Ow, M.C.; Vasquez-Rifo, A.; Rondeau, E.L.; Ambros, V.R.; Großhans, H.; Simard, M.J. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell 2013, 50, 281–287. [Google Scholar] [CrossRef] [Green Version]
- Meziane, O.; Piquet, S.; Bossé, G.D.; Gagné, D.; Paquet, E.; Robert, C.; Tones, M.A.; Simard, M.J. The human decapping scavenger enzyme DcpS modulates microRNA turnover. Sci. Rep. 2015, 5, 16688. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef] [Green Version]
- Pauley, K.M.; Eystathioy, T.; Jakymiw, A.; Hamel, J.C.; Fritzler, M.J.; Chan, E.K. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006, 7, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Detzer, A.; Engel, C.; Wunsche, W.; Sczakiel, G. Cell stress is related to re-localization of Argonaute 2 and to decreased RNA interference in human cells. Nucleic Acids Res. 2011, 39, 2727–2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Chang, L.; Wang, H.; Su, A.; Wu, Z. Dcp1a and GW182 induce distinct cellular aggregates and have different effects on microRNA pathway. DNA Cell Biol. 2017, 36, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.S.; Rüegger, S.; Gaidatzis, D.; Stadler, M.B.; Großhans, H. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 2014, 42, 4056–4067. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. FASTP: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Hu, F.; Sung, M.W.; Shu, C.; Castillo-González, C.; Koiwa, H.; Tang, G.; Dickman, M.; Li, P.; Zhang, X. RISC-interacting clearing 3′-5′exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife 2017, 6, e24466. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. Cell Mol. Biol. 2006, 45, 616–629. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Chang, I.F.; Gong, F.; Galbraith, D.W.; Bailey-Serres, J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005, 138, 624–635. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Niu, D.; Carbonell, A.; Wang, A.; Lee, A.; Tun, V.; Wang, Z.; Carrington, J.C.; Chang, C.E.; Jin, H. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014, 5, 5468. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Gao, W.; Wu, S.; Lu, L.; Chen, Y.; Guo, J.; Men, S.; Zhang, X. AtXRN4 Affects the Turnover of Chosen miRNA*s in Arabidopsis. Plants 2020, 9, 362. https://doi.org/10.3390/plants9030362
Liu Y, Gao W, Wu S, Lu L, Chen Y, Guo J, Men S, Zhang X. AtXRN4 Affects the Turnover of Chosen miRNA*s in Arabidopsis. Plants. 2020; 9(3):362. https://doi.org/10.3390/plants9030362
Chicago/Turabian StyleLiu, Yan, Wenrui Gao, Shuangyang Wu, Lu Lu, Yaqiu Chen, Junliang Guo, Shuzhen Men, and Xiaoming Zhang. 2020. "AtXRN4 Affects the Turnover of Chosen miRNA*s in Arabidopsis" Plants 9, no. 3: 362. https://doi.org/10.3390/plants9030362




