Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia
Abstract
1. Introduction
2. Results
2.1. Genetic Diversity of Olive Genotypes
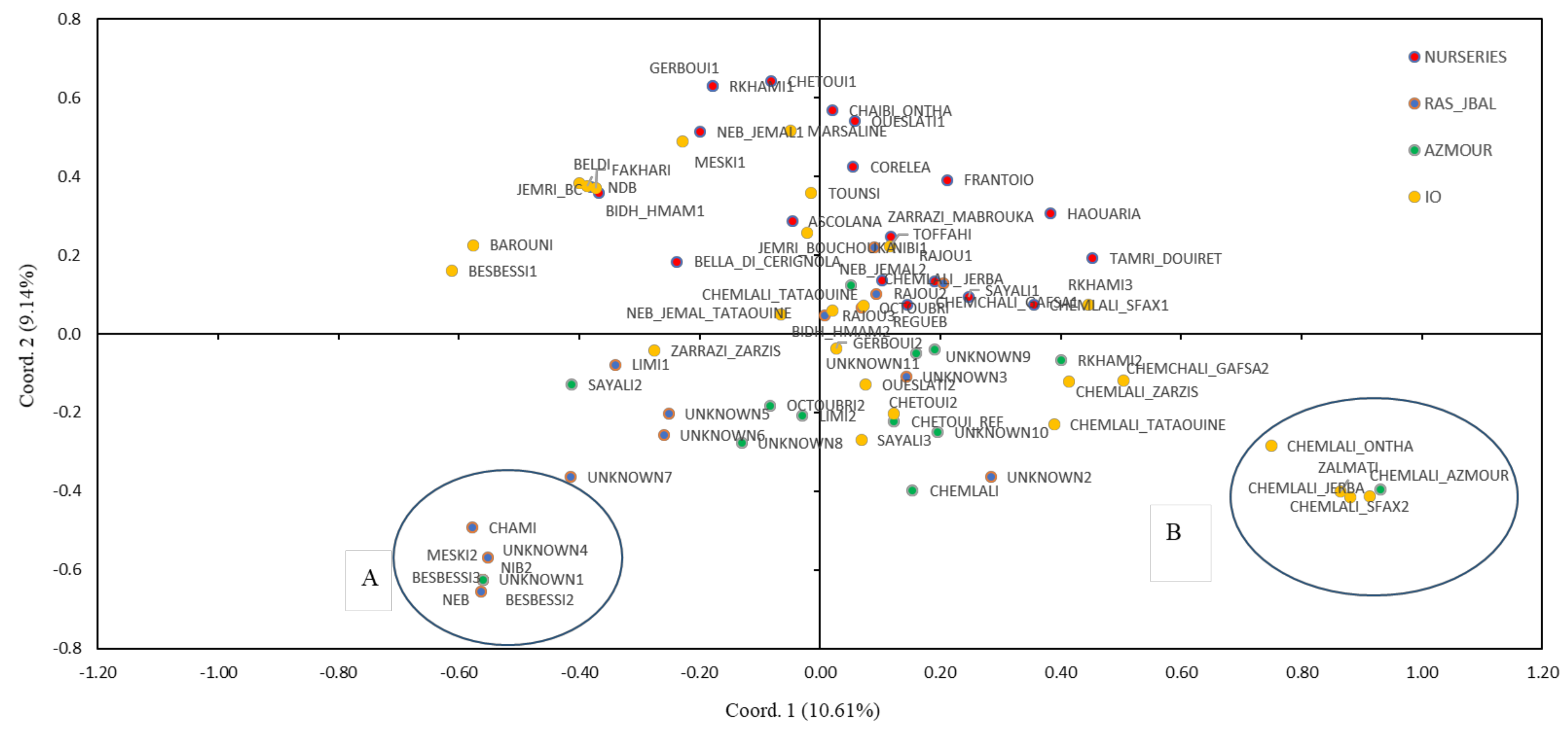
2.2. Genetic Relationships Among Olive Genotypes
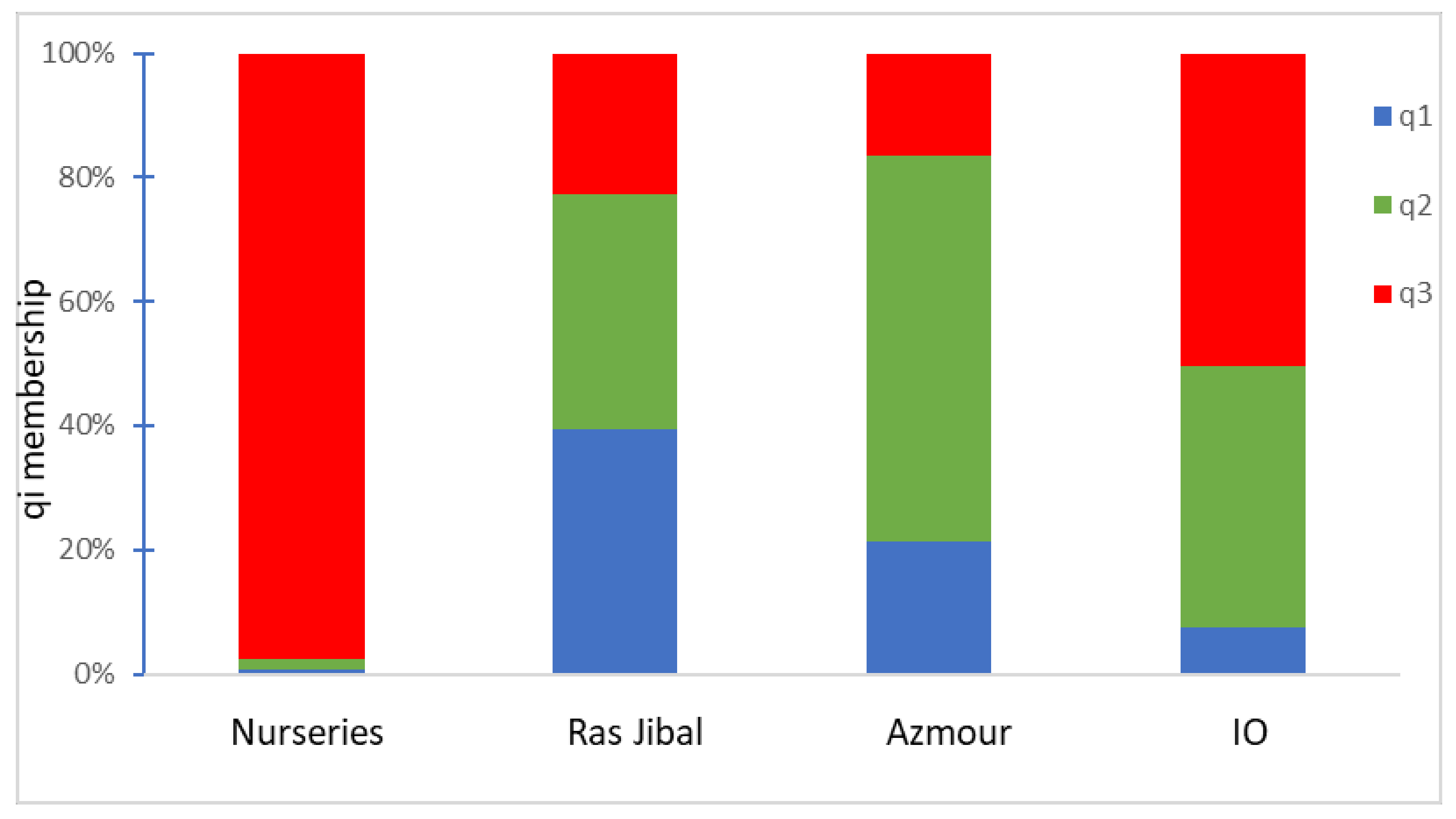
2.3. Genetic Structure
3. Discussion
4. Materials and Methods
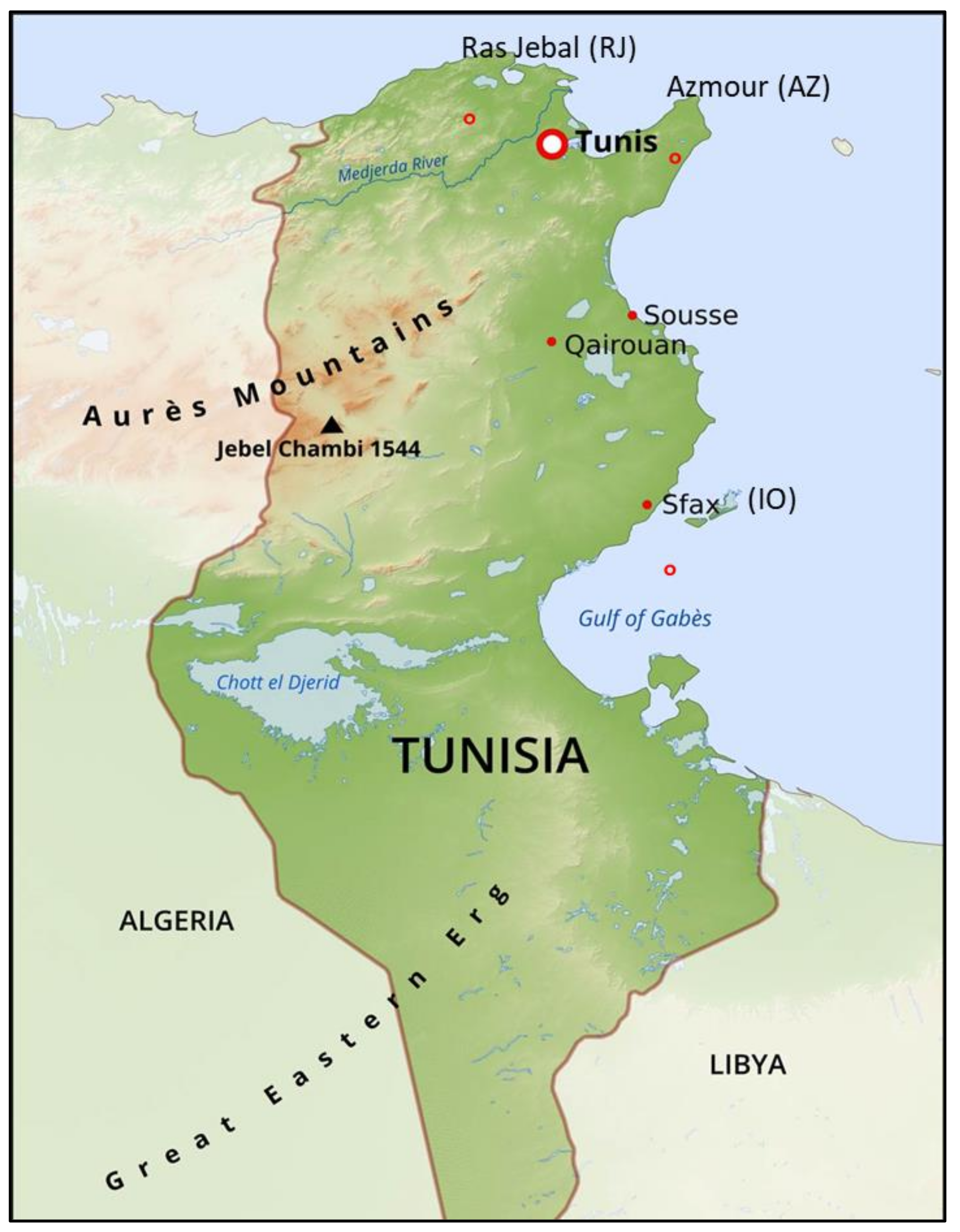
4.1. Plant Material
4.2. DNA Extraction
4.3. SSR Assays
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Faostat. 2017. Available online: http://www.fao.org/faostat/en/?#data/QC (accessed on 20 September 2019).
- Rugini, E.; Baldoni, L.; Muleo, R.; Sebastiani, L. The olive tree genome. In Compendium of Plant Genomes; Rugini, E., Baldoni, L., Muleo, R., Sebastiani, L., Eds.; Springer: Basel, Switzerland, 2017; pp. 27–54. [Google Scholar] [CrossRef]
- Chazan-Gillig, S. Civilisation de l’olivier et des céréales. La Méditerranée assassinée. Peuples Méditerranéens 1993, 62–63, 97–113. [Google Scholar]
- Diez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Besnard, G.; El Bakkali, A.; Haouane, H.; Baali-Cherif, D.; Moukhli, A.; Khadari, B. Population genetics of Mediterranean and Saharan olives: Geographic patterns of differentiation and evidence for early-generations of admixture. Ann. Bot. 2013, 112, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- di Rienzo, V.; Sion, S.; Taranto, F.; D’Agostino, N.; Montemurro, C.; Fanelli, V.; Sabetta, W.; Boucheffa, S.; Tamendjari, A.; Pasqualone, A.; et al. Genetic flow among olive population within the Mediterranean basin. Peer J. 2018, 6. [Google Scholar] [CrossRef]
- Tunisian Agriculture Ministry-General Direction of Agricultural Production. Available online: http://www.agriculture.tn (accessed on 20 September 2019).
- Trigui, A.; Msallem, M. Oliviers de Tunisie: catalogue des Variétés Autochtones & Types Locaux: Identification Variétale and Caractérisation Morpho-pomologique des Ressources Génétiques Oléicoles de Tunisie (vol.1); Institution de la Recherche et de l’Enseignement Supérieur Agricoles: Tunis, Tunisia, 2002; p. 159. [Google Scholar]
- Abaza, L.; Msallem, M.; Daoud, D.; Zarrouk, M. Caractérisation des huiles de sept variétés d’olivier Tunisiennes. Oléagineux Corps Gras Lipides 2002, 9, 174–179. [Google Scholar] [CrossRef][Green Version]
- Taamalli, W.; Geuna, F.; Banfi, R.; Bassi, D.; Daoud, D.; Zarrouk, M. Agronomic and molecular analyses for the characterization of accessions in Tunisian olive germplasm collections. Electron. J. Biotech. 2006, 9, 467–481. [Google Scholar] [CrossRef]
- Abdelhamid, S.; Grati-Kamoun, N.; Marra, F.; Caruso, T. Genetic similarity among Tunisian cultivated olive estimated through SSR markers. Sci. Agric. 2013, 70, 33–38. [Google Scholar] [CrossRef][Green Version]
- Ben Ayed, R.; Kallel, I.; Ben Hassen, H.; Rebai, A. SNP marker analysis for validating the authenticity of Tunisian olive oil. J. Genet. 2014, 93, 48–54. [Google Scholar]
- Abdelhamid, S.; Omri, A.; Grati-Kamoun, N.; Marra, F.P. Molecular characterization and genetic relationships of cultivated Tunisian olive varieties (Olea europaea L.) using SSR markers. J. New Sci. Agric. Biotech. 2017, 40, 2175–2185. [Google Scholar]
- Ben Mohamed, M.; Zelasco, S.; Ben Ali, S.; Guasmi, F.; Triki, T.; Conforti, F.L.; Kamoun Naziha, G. Exploring olive trees genetic variability in the South East of Tunisia. Genet. Mol. Res. 2017, 16, gmr16039850. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Ennouri, K.; Ben Hassen, H.; Triki, M.A.; Rebai, A. Comparison between DNA-based, pomological, and chemical markers accomplished by bioinformatic tools to distinguish within Tunisian olive cultivars. J. Fund. Appl. Sci. 2015, 7, 408–421. [Google Scholar] [CrossRef]
- Abdelhamid, S.; Grati-kamoun, N.; Marra, F.P.; Caruso, T. Genetic characterization of Tunisian Olive Table Cultivars (Olea europaea L.): Inventory Based on Microsatellite Analysis. Afr. J. Plant Sci. Biotechnol. 2010, 4, 3–16. [Google Scholar]
- Taamalli, W.; Geuna, F.; Bassi, D.; Daoud, D. SSR Marker Based DNA Fingerprinting of Tunisian Olive (Olea europaea L.) Varieties. J. Agric. 2008, 7, 176–181. [Google Scholar]
- Ben Mohamed, M.; Ben Ali, S.; Boussora, F.; Guasmi, F.; Triki, T. Polymorphism of Microsatellite (SSR) Markers in Tunisian Olive (Olea europaea L.) Cultivars. J. Mult. Engin. Sc. St. 2017, 3, 1247–1252. [Google Scholar]
- Mazzeo, R.; Morgese, A.; Sonnante, G.; Zualuaga, D.; Pavan, S.; Ricciardi, L.; Lotti, C. Genetic Diversity in broccoli rabe (Brassica rapa L. subsp. sylvestris (L.) Janch.) from Southern Italy. Sci. Hortic. 2019, 253, 140–146. [Google Scholar]
- Taranto, F.; Francese, G.; Di Dato, F.; D’Alessandro, A.; Greco, B.; Onofaro Sanajà, V.; Pentangelo, A.; Mennella, G.; Tripodi, P. Leaf metabolic, genetic, and morphophysiological profiles of cultivated and wild rocket salad (Eruca and Diplotaxis spp.). J. Agric. Food Chem. 2016, 64, 5824–5836. [Google Scholar] [CrossRef]
- De Giovanni, C.; Pavan, S.; Taranto, F.; Di Rienzo, V.; Miazzi, M.M.; Marcotrigiano, A.R.; Mangini, G.; Ricciardi, L.; Lotti, C. Genetic variation of a global germplasm collection of chickpea (Cicer arietinum L.) including Italian accessions at risk of genetic erosion. Physiol. Mol. Biol. Plants 2017, 23, 197–205. [Google Scholar] [CrossRef]
- Fanelli, V.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Montemurro, C.; La Notte, P.; Miazzi, M.M.; Bruno, M.; Falbo, M.; Petrillo, F.; et al. Molecular characterization of wine grape cultivars from Calabria. Acta Hortic. 2019, 1248, 281–286. [Google Scholar] [CrossRef]
- di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Sabetta, W.; Montemurro, C. The preservation and characterization of Apulian olive germplasm biodiversity. Acta Hortic. 2018, 1199, 1–6. [Google Scholar] [CrossRef]
- Muzzalupo, I.; Vendramin, G.G.; Chiappetta, A. Genetic biodiversity of Italian olives (Olea europaea) germplasm analyzed by SSR markers. Sci. World J. 2014, 2014, 296590. [Google Scholar] [CrossRef]
- Veloso, M.M.; Simões-Costa, M.C.; Carneiro, L.C.; Guimarães, J.B.; Mateus, C.; Fevereiro, P.; Pinto-Ricardo, C. Olive Tree (Olea europaea L.) Diversity in Traditional Small Farms of Ficalho; Portugal. Diversity 2018, 10, 5. [Google Scholar] [CrossRef]
- Montemurro, C.; Miazzi, M.M.; Pasqualone, A.; Fanelli, V.; Sabetta, W.; Di Rienzo, V. Traceability of PDO olive oil “Terra di Bari” using high resolution melting. J. Chem. 2015, 2015, 496986. [Google Scholar] [CrossRef]
- Pasqualone, A.; di Rienzo, V.D.; Miazzi, M.M.; Fanelli, V.; Caponio, F.; Montemurro, C. High resolution melting analysis of DNA microsatellites in olive pastes and virgin olive oils obtained by talc addition. Eur. J. Lipid Sci. Technol. 2015, 117, 2044–2048. [Google Scholar] [CrossRef]
- Sabetta, W.; Miazzi, M.M.; di Rienzo, V.; Fanelli, V.; Pasqualone, A.; Montemurro, C. Development and application of protocols to certify the authenticity and the traceability of Apulian typical products in olive sector. Rivista Italiana Delle Sostanze Grasse 2017, 94, 37–43. [Google Scholar]
- Di Rienzo, V.; Fanelli, V.; Miazzi, M.M.; Sabetta, W.; Montemurro, C. A reliable analytical procedure to discover table grape DNA adulteration in industrial wines and musts. Acta Hortic. 2017, 1188, 365–370. [Google Scholar] [CrossRef]
- Binetti, G.; Del Coco, L.; Ragone, R.; Zelasco, S.; Perri, E.; Montemurro, C.; Valentini, R.; Naso, D.; Fanizzi, F.P.; Schena, F.P. Cultivar classification of Apulian olive oils: Use of artificial neural networks for comparing NMR, NIR and merceological data. Food Chem. 2017, 219, 131–138. [Google Scholar] [CrossRef]
- Abdelhamid, S.; Gouta, H.; Gharsallaoui, M.; Ghrab, M.; Kwon, Y.T.; Yoon, I.S.; Byun, M.O. A Review on Current Status of Olive and Olive Oil Production in Tunisia. J. Korean Soc. Int. Agric. 2013, 25, 351–357. [Google Scholar] [CrossRef]
- Muzzalupo, I.; Muto, A.; Badolati, G.; Veizi, A.; Chiappetta, A. Genotyping of Albania olive (Olea europaea) germplasm by SSR molecular markers. Emir. J. Food Agric. 2018, 30, 573–580. [Google Scholar] [CrossRef]
- Mnasri, S.; Saddoud Debbabi, O.; M’barek, B.N. Molecular markers: An important tool to analyze the genetic diversity of local Tunisian olive varieties. Euro-Medit. J. Environ. Integr. 2019, 4, 29. [Google Scholar] [CrossRef]
- Grati Kamoun, N.; Khlif, M. Caracterisation technologique des varietes d’olivier cultivees en Tunisie. Ezzaitouna 2001, 69, 1892–1900. [Google Scholar]
- Rekik, I.; Salimonti, A.; Grati Kamoun, N.; Muzzalupo, I.; Lepais, O.; Gerber, S.; Perri, E.; Rebai, A. Characterization and Identification of Tunisian Olive Tree Varieties by Microsatellite Markers. HortScience 2008, 43, 1371–1376. [Google Scholar] [CrossRef]
- Boucheffa, S.; Miazzi, M.M.; di Rienzo, V.; Mangini, G.; Fanelli, V.; Tamendjari, A.; Pignone, D.; Montemurro, C. The coexistence of oleaster and traditional varieties affects genetic diversity and population structure in Algerian olive (Olea europaea) germplasm. Genet. Res. Crop Evol. 2017, 64, 379–390. [Google Scholar] [CrossRef]
- Boucheffa, S.; Tamendjari, A.; Sanchez-Gimeno, A.C.; Rovellini, P.; Venturini, S.; di Rienzo, V.; Miazzi, M.M.; Montemurro, C. Diversity Assessment of Algerian Wild and Cultivated Olives (Olea europaea L.) by Molecular; Morphological and Chemical Traits. Eur. J. Lipid Sci. Technol. 2019, 121, 1800302. [Google Scholar] [CrossRef]
- Babay, E.; Khamassi, K.; Sabetta, W.; Miazzi, M.M.; Montemurro, C.; Pignone, D.; Danzi, D.; Finetti Sialer, M.M.; Mangini, G. Serendipitous in situ Conservation of Faba Bean Landraces in Tunisia: A Case Study. Genes 2020, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R. Sanitary and phytosanitary (SPS) and related non-tariff barriers to agricultural trade. In Agricultural Trade: Sanitary, Phytosanitary and Technical Barriers. 2014. Available online: www.crs (accessed on 19 March 2019).
- Ben Ayed, R.; Ben Hassen, H.; Ennouri, K.; Ben Marzoug, R.; Rebai, A. OGDD (Olive Genetic Diversity Database): A microsatellite markers’ genotypes database of worldwide olive trees for cultivar identification and virgin olive oil traceability. Database 2016. [Google Scholar] [CrossRef] [PubMed]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Saddoud Debbabi, O.; Ben Amar, F.; et al. Genetic Characterization of Apulian Olive Germplasm as Potential Source in New Breeding Programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G. A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Caruso, T.; Marra, F.P.; Costa, F.; Campisi, G.; Macaluso, L.; Marchese, A. Genetic diversity and clonal variation within the main Sicilian olive cultivars based on morphological traits and microsatellite markers. Sci. Hortic. 2014, 180, 130–138. [Google Scholar] [CrossRef]
- Belaj, A.; del Carmen Dominguez-García, M.; Atienza, S.G.; Urdíroz, N.M.; De la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V.; et al. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Hannachi, H.; Breton, C.; Msallem, M.; El Hadj, S.B.; El Gazzah, M.; Bervillé, A. Genetic Relationships between Cultivated and Wild Olive Trees (Olea europaea L. var europaea and var. sylvestris) Based on Nuclear and Chloroplast SSR Markers. Nat. Res. 2010, 1, 95–103. [Google Scholar] [CrossRef]
- Khadari, B.; Charafi, J.; Moukhli, A.; Ater, M. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: A paradoxical situation evidenced by the use of SSR loci. Tree Genet. Genomes 2008, 4, 213–221. [Google Scholar] [CrossRef]
- Yoruk, B.; Tuskin, V. Genetic diversity and relationships of wild and cultivated olives in Turkey. Plant Syst. Evol. 2014, 300, 1247–1258. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Ennouri, K.; Ben Hassen, H.; Rebai, A. Molecular phylogeny to specify Zalmati and Chemlali Tataouine Tunisian olive cultivars. J. New Sci. 2015, 8, 689–694. [Google Scholar]
- D’Agostino, N.; Taranto, F.; Camposeo, S.; Mangini, G.; Fanelli, V.; Gadaleta, S.; Miazzi, M.M.; Pavan, S.; Di Rienzo, V.; Sabetta, W.; et al. GBS-derived SNP catalogue unveiled wide genetic variability and geographical relationships of Italian olive cultivars. Sci. Rep. 2018, 8, 15877. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Fanelli, V.; di Rienzo, V.; Miazzi, M.M.; Pavan, S.; Zelasco, S.; Perri, E.; Montemurro, C. SNP diversity in an olive germplasm collection. Acta Hortic. 2018, 1199, 27–32. [Google Scholar] [CrossRef]
- Barabaschi, D.; Tondelli, A.; Desiderio, F.; Volante, A.; Vaccino, P.; Valè, G.; Cattivelli, L. Next generation breeding. Plant Sci. 2016, 242, 3–13. [Google Scholar] [CrossRef]
- Belaj, A.; De la Rosa, R.; Lorite, I.J.; Mariotti, R.; Cultrera, N.G.M.; Beuzón, C.R.; González-Plaza, J.J.; Muñoz-Mérida, A.; Trelles, O.; Baldoni, L. Usefulness of a New Large Set of High Throughput EST-SNP Markers as a Tool for Olive Germplasm Collection Management. Front. Plant Sci. 2018, 9, 215. [Google Scholar] [CrossRef]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, D.; Miazzi, M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 2019, 21, 1215–1226. [Google Scholar]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Santos, M.R.D.; Machado, M.L.D.C.; Machado, A.D.C. Identification of microsatellite loci in olive (Olea europaea L.) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef]
- Cipriani, G.; Marrazzo, M.T.; Marconi, R.; Cimato, A.; Testolin, R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual finger-printing and reveal polymorphism within ancient cultivars. Theor. Appl. Genet. 2002, 104, 223–228. [Google Scholar] [CrossRef]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E.; Weaver, W. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of Pairwise Relatedness with Molecular Markers. Genetics 1999, 152, 1753–1766. [Google Scholar]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST). Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef] [PubMed]


| Locus | Size Range (bp) | Na | Ne | I | Ho | He | F | PIC |
|---|---|---|---|---|---|---|---|---|
| DCA03 | 231–255 | 10 | 5.89 | 1.91 | 0.92 | 0.83 | −0.10 | 0.80 |
| DCA05 | 194–212 | 7 | 3.04 | 1.44 | 0.77 | 0.67 | −0.16 | 0.64 |
| DCA09 | 162–206 | 13 | 5.94 | 2.09 | 0.85 | 0.83 | −0.03 | 0.81 |
| DCA15 | 246–270 | 4 | 1.84 | 0.88 | 0.39 | 0.45 | 0.14 | 0.42 |
| DCA16 | 122–186 | 19 | 9.26 | 2.51 | 0.97 | 0.89 | −0.09 | 0.88 |
| DCA17 | 109–181 | 9 | 3.11 | 1.39 | 0.25 | 0.67 | 0.62 | 0.63 |
| DCA18 | 165–191 | 10 | 4.76 | 1.76 | 0.61 | 0.79 | 0.21 | 0.76 |
| GAPU71b | 121–144 | 5 | 4.91 | 1.60 | 0.89 | 0.79 | −0.12 | 0.76 |
| GAPU101 | 170–218 | 9 | 6.75 | 1.96 | 0.97 | 0.85 | −0.14 | 0.83 |
| UDO28 | 115–169 | 17 | 6.77 | 2.20 | 0.79 | 0.85 | 0.07 | 0.83 |
| UDO43 | 166–216 | 15 | 7.03 | 2.19 | 0.90 | 0.85 | −0.06 | 0.84 |
| EMOL | 190–228 | 6 | 2.70 | 1.18 | 0.45 | 0.63 | 0.27 | 0.57 |
| Total | 124 | 62.00 | ||||||
| Mean | 10.33 | 5.16 | 1.76 | 0.73 | 0.76 | 0.05 | 0.73 | |
| Collections | Na | Ne | Ho | He | F | |
|---|---|---|---|---|---|---|
| Reference (IO) | Total | 92.0 | 55.7 | |||
| Mean | 6.4 | 4.21 | 0.67 | 0.70 | 0.078 | |
| Nurseries (GR) | Total | 77.0 | 50.5 | |||
| Mean | 6.4 | 4.21 | 0.67 | 0.70 | 0.078 | |
| Ras Jbal (RJ) | Total | 74.0 | 44.5 | |||
| Mean | 6.1 | 3.71 | 0.82 | 0.67 | −0.219 | |
| Azmour (AZ) | Total | 82.0 | 55.5 | |||
| Mean | 6.8 | 4.63 | 0.79 | 0.73 | −0.066 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saddoud Debbabi, O.; Miazzi, M.M.; Elloumi, O.; Fendri, M.; Ben Amar, F.; Savoia, M.; Sion, S.; Souabni, H.; Mnasri, S.R.; Ben Abdelaali, S.; et al. Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia. Plants 2020, 9, 382. https://doi.org/10.3390/plants9030382
Saddoud Debbabi O, Miazzi MM, Elloumi O, Fendri M, Ben Amar F, Savoia M, Sion S, Souabni H, Mnasri SR, Ben Abdelaali S, et al. Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia. Plants. 2020; 9(3):382. https://doi.org/10.3390/plants9030382
Chicago/Turabian StyleSaddoud Debbabi, Olfa, Monica Marilena Miazzi, Olfa Elloumi, Mahdi Fendri, Fathi Ben Amar, Michele Savoia, Sara Sion, Hana Souabni, Sameh Rahmani Mnasri, Selma Ben Abdelaali, and et al. 2020. "Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia" Plants 9, no. 3: 382. https://doi.org/10.3390/plants9030382
APA StyleSaddoud Debbabi, O., Miazzi, M. M., Elloumi, O., Fendri, M., Ben Amar, F., Savoia, M., Sion, S., Souabni, H., Mnasri, S. R., Ben Abdelaali, S., Jendoubi, F., Mangini, G., Famiani, F., Taranto, F., Montemurro, C., & Msallem, M. (2020). Recovery, Assessment, and Molecular Characterization of Minor Olive Genotypes in Tunisia. Plants, 9(3), 382. https://doi.org/10.3390/plants9030382











