Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca acinosa Roxb
Abstract
:1. Introduction
2. Results
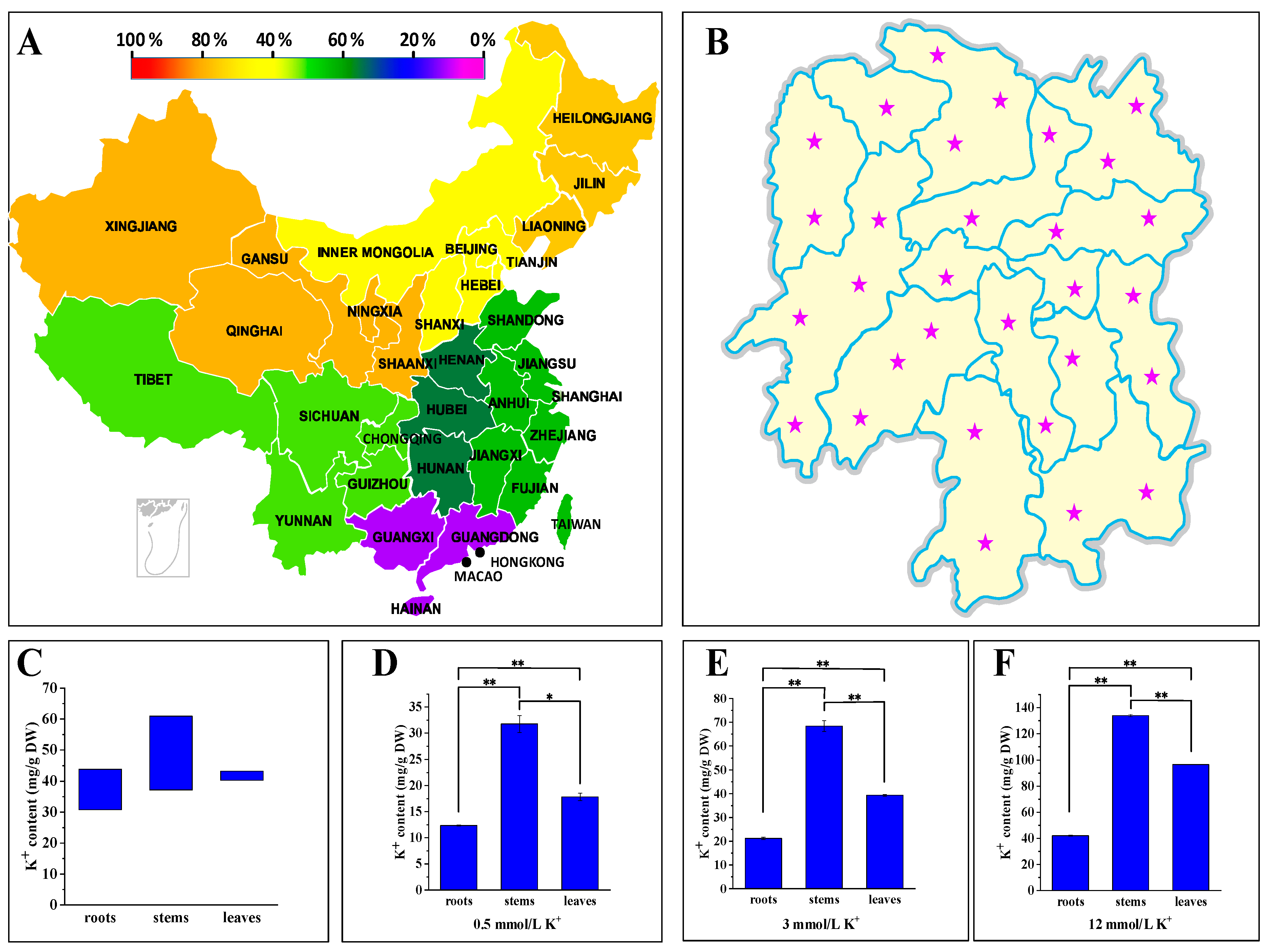
2.1. P. acinosa Showed High Potassium Characteristics
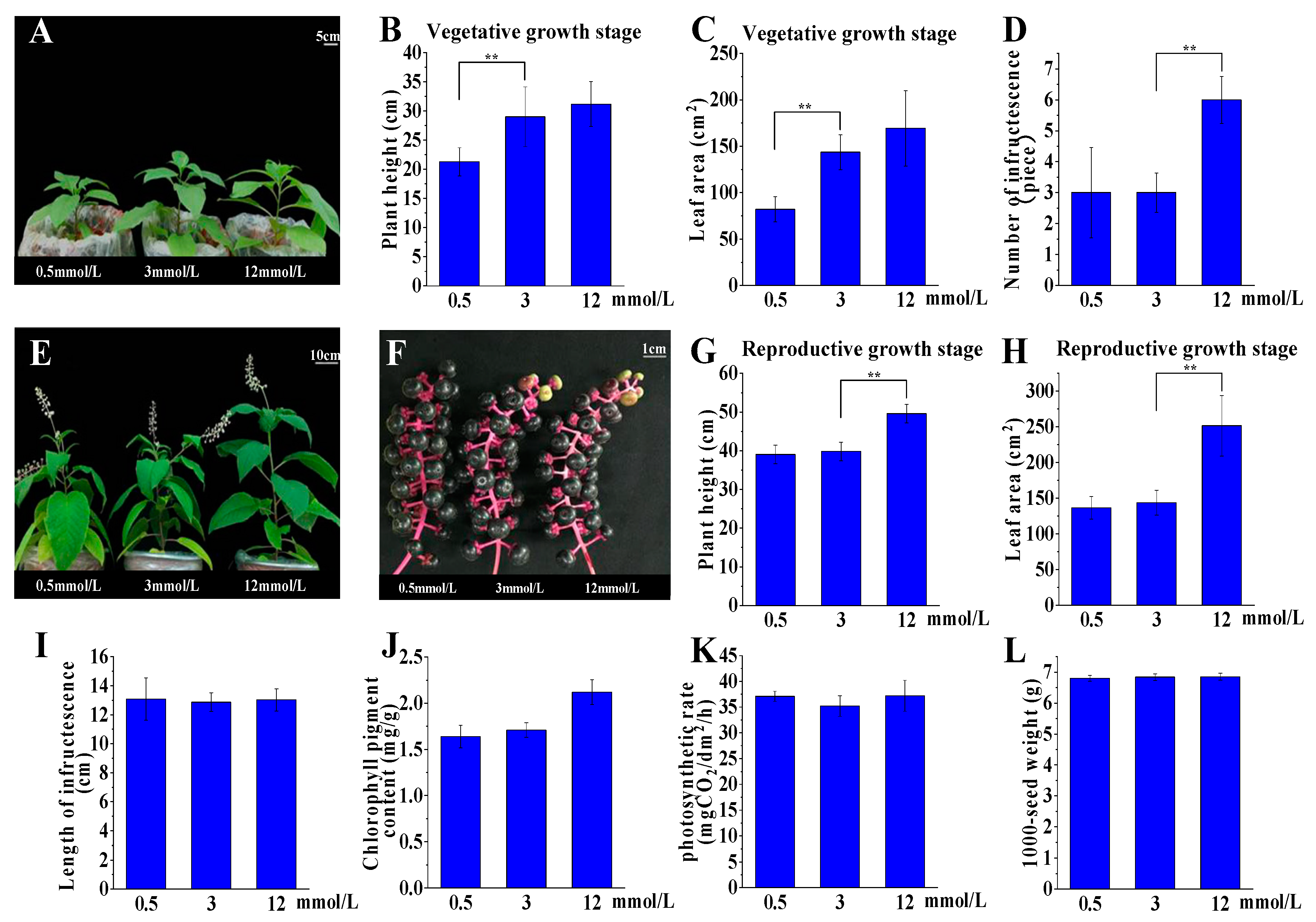
2.2. P. acinosa Shows Adaptability to a Wide Range of External Potassium Levels
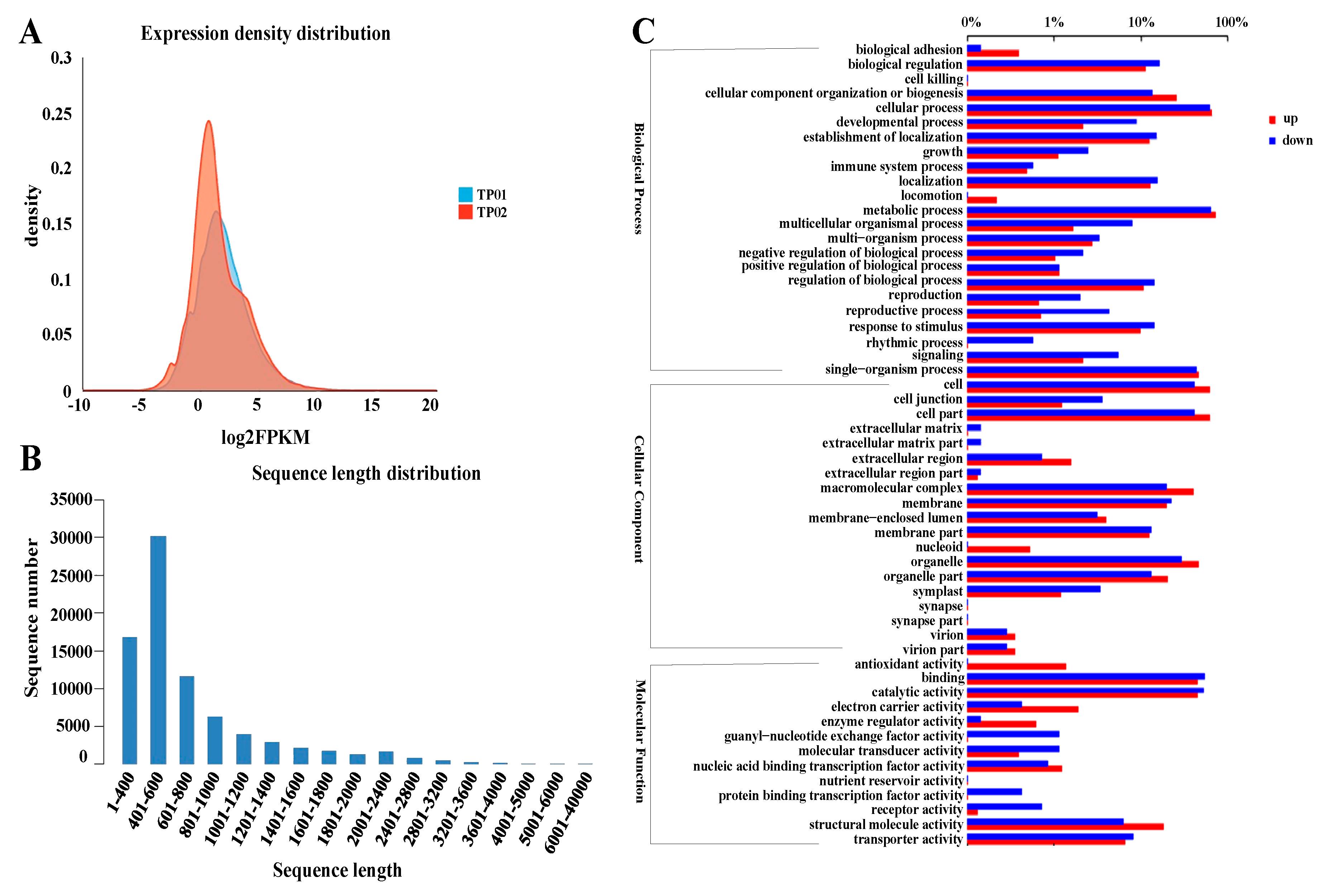
2.3. Global Transcriptome Analysis Reveals the Involvement of Multiple Biological Processes during Potassium Accumulation
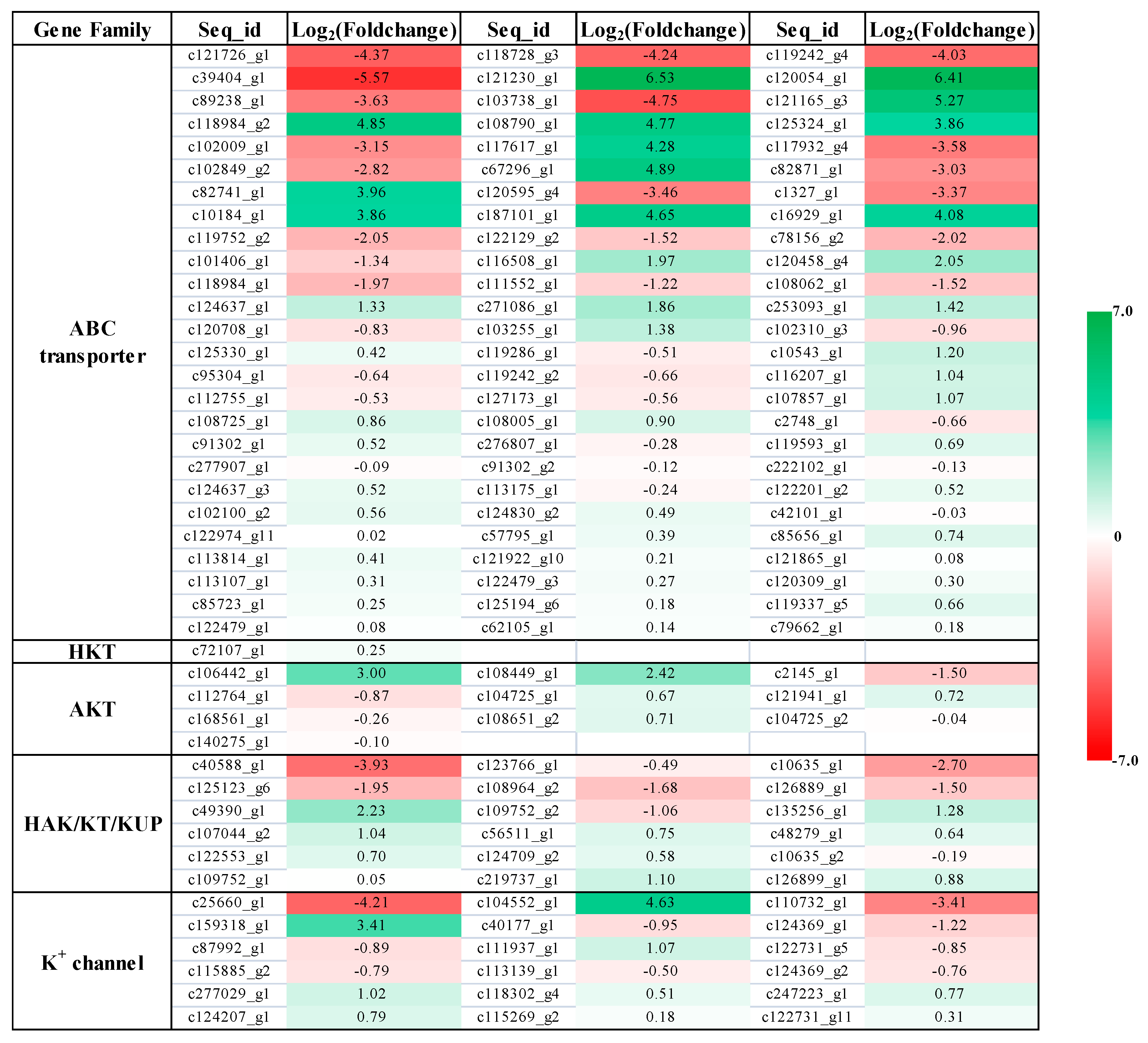
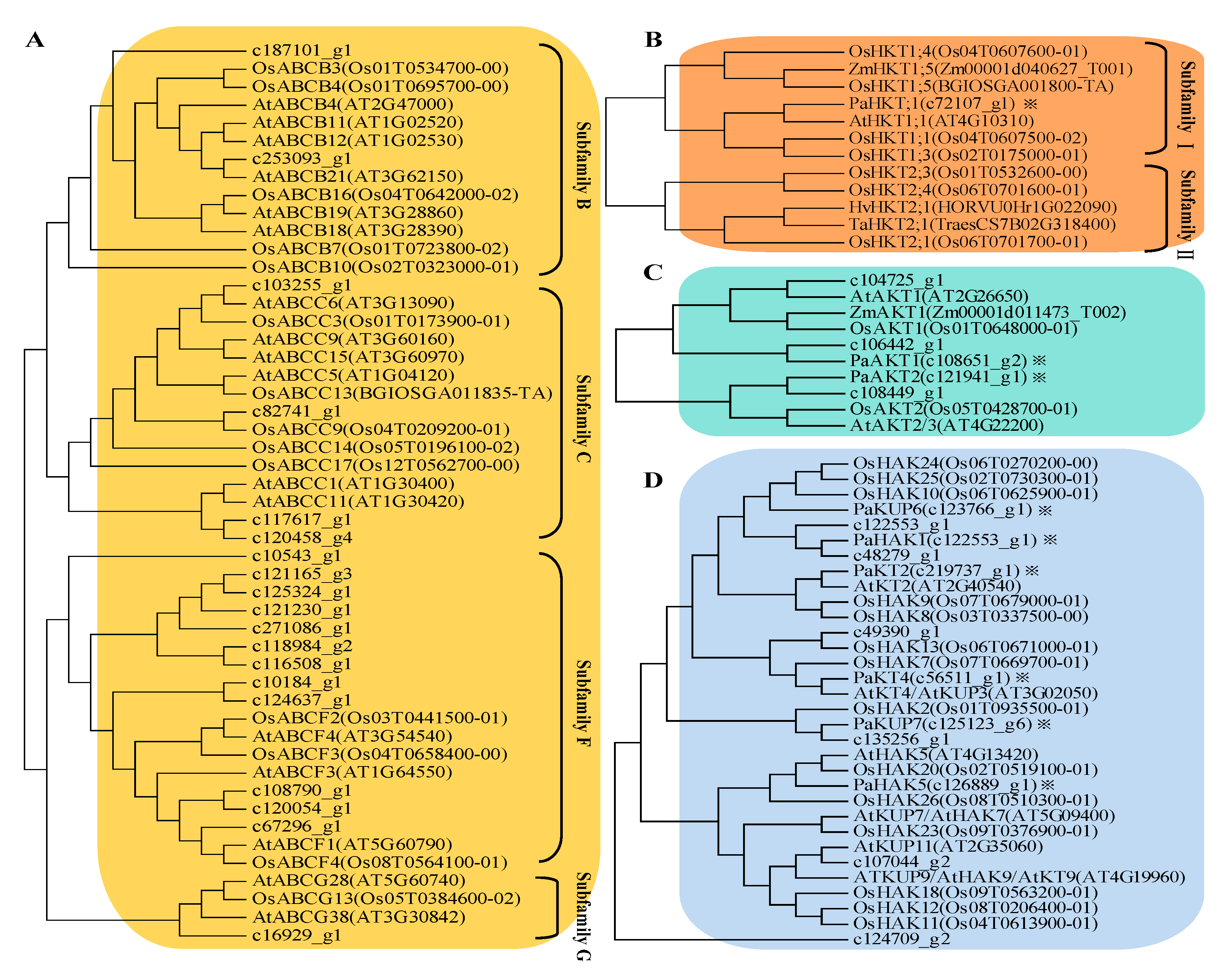
2.4. Expression Profile of K+ Transporters Based on Transcriptome Sequence
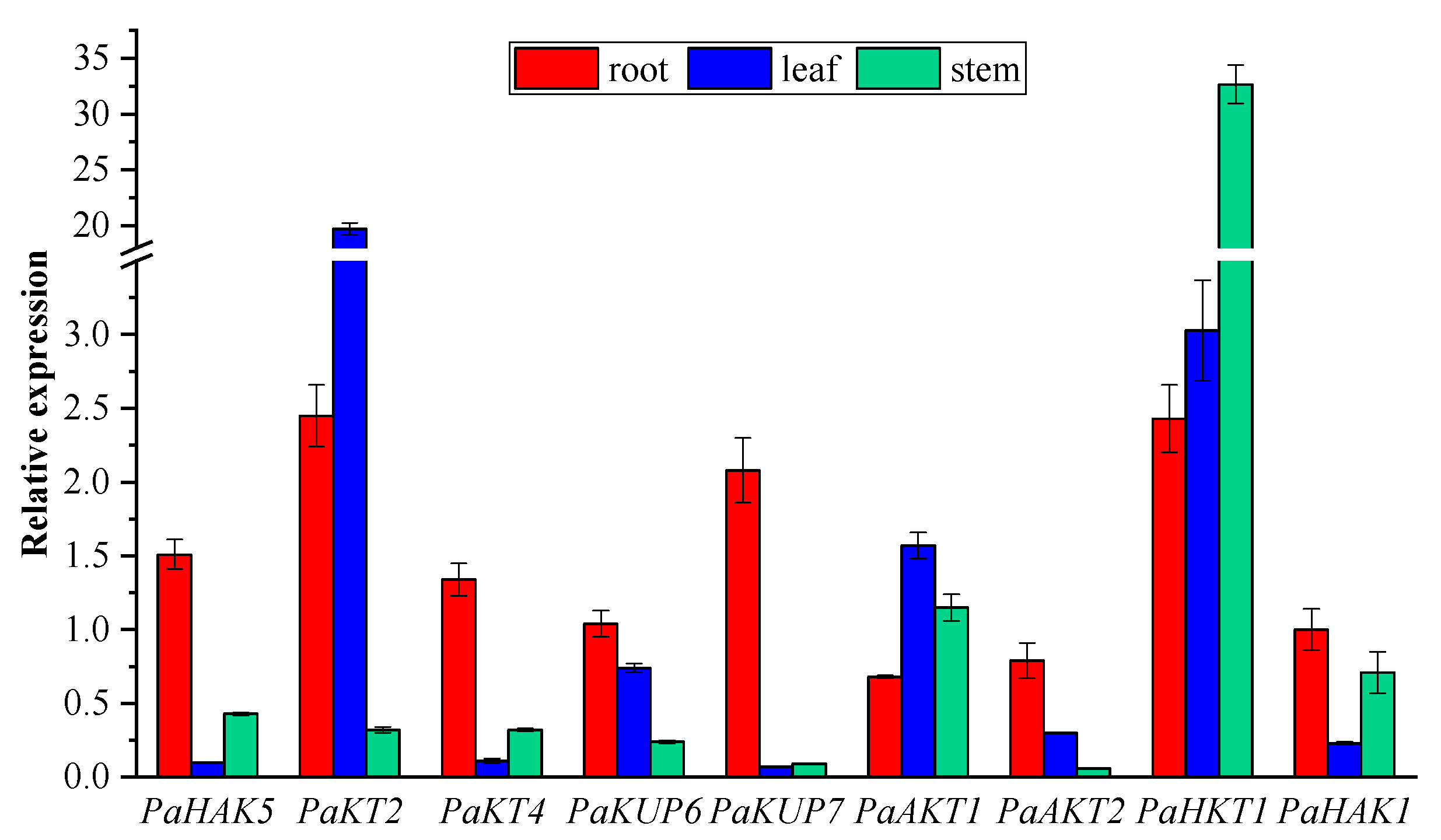
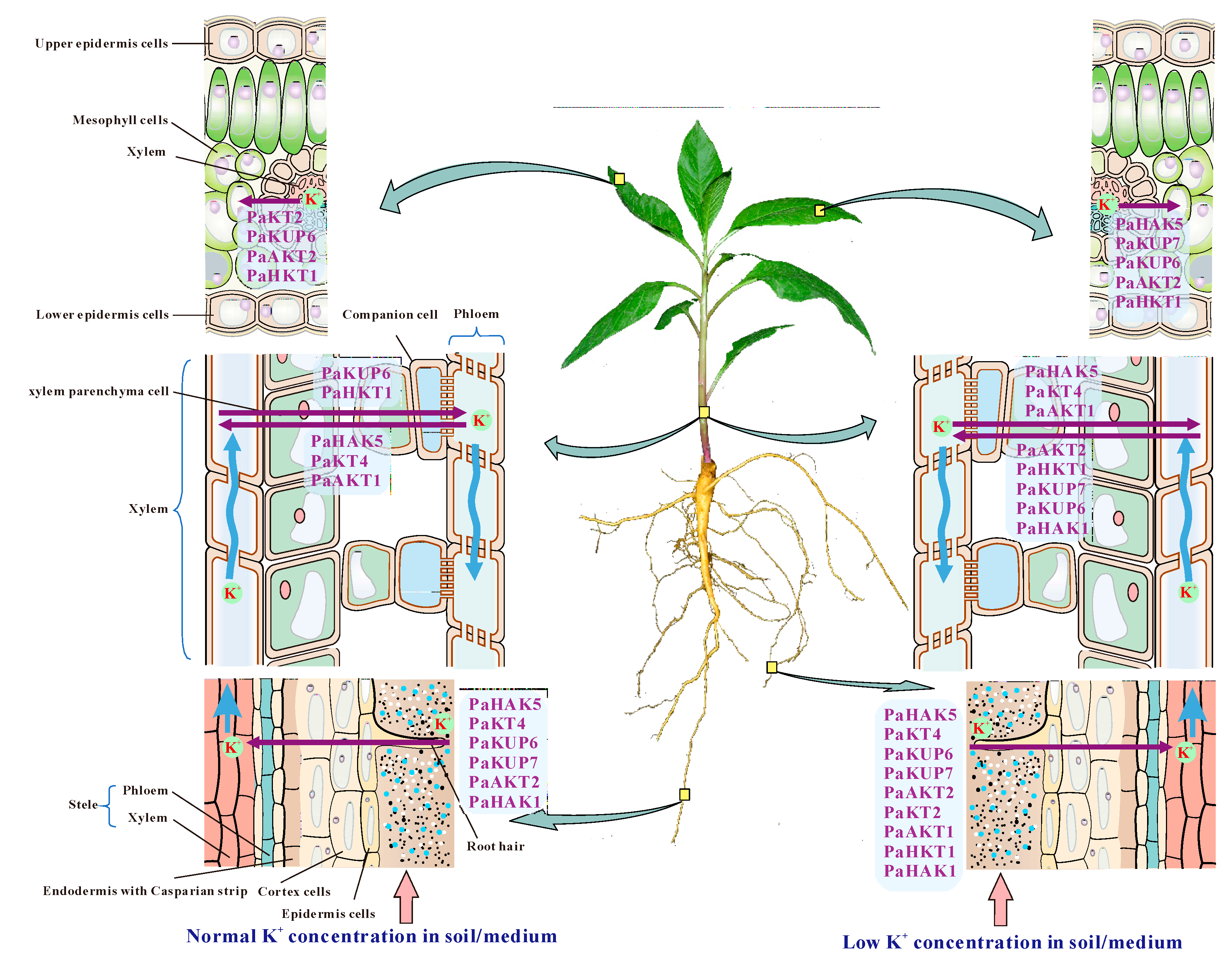
2.5. Expression Pattern of Potassium Transport Genes in P. acinosa
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Collection from Fields
5.2. Plant Materials and Growth Conditions
5.3. K+ Treatment for Physiological Tests
5.4. K+ Treatment for Transcriptome Sequencing
5.5. Measurement of Chlorophyll Content and Photosynthetic Rate in Leaves
5.6. K+ Content Analysis through ICP–MS
5.7. Transcriptome Analysis
5.8. Real-Time Quantitative PCR Analysis
5.9. Gene Searching and Phylogenetic Analysis
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amtmann, A.; Blatt, M.R. Regulation of macronutrient transport. New Phytol. 2009, 181, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Gajdanowicz, P.; Michard, E.; Sandmann, M.; Rocha, M.; Corrêa, L.G.G.; Ramírez-Aguilar, S.J.; Gomez-Porras, J.L.; González, W.; Thibaud, J.B.; Van Dongen, J.T.; et al. Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc. Natl. Acad. Sci. USA 2011, 108, 864–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, R.A.; Wyn, J. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Anschutz, U.; Becker, D.; Shabala, S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef]
- Song, Z.B.; Wu, X.F.; Gao, Y.L.; Cui, X.; Jiao, F.C.; Chen, X.J.; Li, Y.P. Genome-wide analysis of the HAK potassium transporter gene family reveals asymmetrical evolution in tobacco (Nicotiana tabacum). Genome 2019, 62, 267–278. [Google Scholar] [CrossRef]
- Li, W.L.; Li, S.Q.; Zhang, Y. A Review on the potassium uptake mechanism in higher plants. J. Maize Sci. 2010, 18, 149–152. [Google Scholar]
- White, P.J.; Earnshaw, M.J.; Clarkson, D.T. Effects of growth and assay temperatures on unidirectional K+ fluxes in roots of rye (Secale cereale). J. Exp. Bot. 1991, 42, 1031–1041. [Google Scholar] [CrossRef]
- Walker, D.J.; Leigh, R.A.; Miller, A.J. Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 1996, 93, 10510–10514. [Google Scholar] [CrossRef] [Green Version]
- Drew, M.C.; Webb, J.; Saker, L.R. Regulation of K+ uptake and transport to the xylem in barley roots, K+ distribution determined by electron probe X-ray microanalysis of frozen-hydrated cells. J. Exp. Bot. 1990, 41, 815–825. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Lara, A.; Ródenas, R.; Jesús, A.; Rubio, F. Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell Environ. 2019, 42, 2357–2371. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. Cellular mechanisms of potassium transport in plants. Physiol. Plant. 2008, 133, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E.; Rains, D.; Elzam, O. Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. USA 1963, 49, 684–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, R.; Schachtman, D.P. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 2004, 101, 8827–8832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maathuis, F.J.M.; Sanders, D. Energization of potassium uptake in Arabidopsis thaliana. Planta 1993, 191, 302–307. [Google Scholar] [CrossRef]
- Wu, S.J.; Ding, L.; Zhu, J.K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Cheng, N.H.; Motes, C.M.; Blancaflor, E.B.; Moore, M.; Gonzales, N.; Padmanaban, S.; Sze, H.; Ward, J.M.; Hirschi, K.D. AtCHX13 is a plasma membrane K+ transporter. Plant Physiol. 2008, 148, 796–807. [Google Scholar] [CrossRef] [Green Version]
- Leidi, E.O.; Barragán, V.; Rubio, L.; El-Hamdaoui, A.; Ruiz, M.T.; Cubero, B.; Ferna´ ndez, J.A.; Bressan, R.A.; Hasegawa, P.M.; Quintero, F.J.; et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010, 61, 495–506. [Google Scholar] [CrossRef] [Green Version]
- Barragán, V.; Leidi, E.O.; Andres, Z.; Rubio, L.; Luca, A.D.; Fernández, J.A.; Cubero, B.; Pardoa, J.M. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [Green Version]
- Reintanz, B.; Szyroki, A.; Ivashikina, N.; Ache, P.; Godde, M.; Becker, D.; Palem, K.; Hedrich, R. AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. USA 2002, 99, 4079–4084. [Google Scholar] [CrossRef] [Green Version]
- Venema, K.; Belver, A.; Marin-Manzano, M.C.; Rodríguez-Rosales, M.P.; Donaire, J.P. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 2003, 278, 22453–22459. [Google Scholar] [CrossRef] [Green Version]
- White, P.; Karley, A. “Potassium”. In Cell Biology of Metals and Nutrients; Hell, R., Mendel, R.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 199–224. [Google Scholar]
- Okada, T.; Nakayama, H.; Shinmyo, A.; Yoshida, K. Expression of OsHAK genes encoding potassium ion transporters inrice. Plant Biotechnol. 2008, 25, 241–245. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gierth, M.; Mäser, P. Potassium transporters in plants–involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007, 581, 348–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, F.; Nieves-Cordones, M.; Alemán, F.; Martínez, V. Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant. 2008, 134, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Gierth, M.; Schroeder, P.M.I. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Hu, Q.D.; Luo, L.; Yang, T.Y.; Zhang, S.; Hu, Y.B.; Xu, G.H. Rice potassium transporter OsHAK1 is essential for maintaining coleoptiles. Russ. J. Plant Physiol. 2015, 42, 231–236. [Google Scholar]
- Yang, T.Y.; Zhang, S.; Hu, Y.B.; Wu, F.C.; Hu, Q.D.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [Green Version]
- Banũelos, M.A.; Garciadeblas, B.; Cubero, B.; Rodríguez-Navarro, A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002, 130, 784–795. [Google Scholar] [CrossRef] [Green Version]
- Benito, B.; Haro, R.; Amtmann, A.; Cuin, T.A.; Dreyer, I. The twins K+ and Na+ in plants. J. Plant Physiol. 2014, 171, 723–731. [Google Scholar] [CrossRef]
- Standard for Nutrient Classification of Soil Census in China. Available online: http://vdb3.soil.csdb.cn/ (accessed on 5 April 2020).
- Su, Y.; Luo, W.G.; Zhao, X.; Ma, L.Y.; Liu, H.Z.; Lin, W.H. Functional analysis of a high-affinity potassium transporter PaHAK1 from P.acinosa by overexpression in eukaryotes. Plant Soil 2015, 397, 63–73. [Google Scholar] [CrossRef]
- Jiang, C.Q.; Zheng, Q.S.; Zu, C.L. Research progress on effects of straw returning on soil potassium and its substitute for potassium fertilizer. Chin. J. Ecol. 2015, 34, 1158–1165. [Google Scholar]
- Hu, D.J.; Yang, M.Y.; Liu, G.H. Studies on high potassium plants. J. Hunan Agric. Univ. (Nat. Sci.) 1980, 4, 8–16. [Google Scholar]
- Tari, M.; Afzal, M.N.; Muhammad, D.; Ahmad, S.; Shahzad, A.N.; Kiran, A.; Wakeel, A. Relationship of tissue potassium content with yield and fiber quality components of Bt cotton as influenced by potassium application methods. Field Crop. Res. 2018, 229, 37–43. [Google Scholar] [CrossRef]
- Clement, C.R.; Hopper, M.J. The supply of potassium to high yielding cut grass. N.A.A.S. Q. Rev. 1968, 79, 101–109. [Google Scholar]
- Hu, D.; Dong, R.; Ge, D. Theory and Practice of Potassium Nutrition in Plant; Hunan Science and Technology Press: Changsha, China, 1993. [Google Scholar]
- Ma, L.Y.; Su, Y.; Guo, Z.Q.; Zhao, X.; Lin, W.H. Characteristics of K+ uptake in Phytolacca Acinosa. J. Hunan Agric. Univ. 2011, 37, 621–623. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC transporters: From microorganisms toman. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef] [PubMed]
- Mengel, K.; Kosegarten, H.; Kirkby, E.A.; Appel, T. Principles of plant nutrition. Springer Sci. Bus. Media 2001. [Google Scholar]
- Sparks, D.L. Potassium dynamics in soils. In Advances in Soil Science; Stewart, B.A., Ed.; Springer: New York, NY, USA, 1987. [Google Scholar]
- Römheld, V.; Kirkby, E.A. Research on potassium in agriculture: Needs and prospects. Plant Soil 2010, 335, 155–180. [Google Scholar] [CrossRef]
- Hirsch, R.E.; Lewis, B.D.; Spalding, E.P.; Sussman, M.R. A role for the AKT1 potassium channel in plant nutrition. Science 1998, 280, 918–921. [Google Scholar] [CrossRef]
- Broadley, M.R.; Escobar-Gutiérrez, A.J.; Bowen, H.C.; Willey, N.J.; White, P.J. Influx and accumulation of Cs+ by the akt1 mutant of Arabidopsis thaliana (L.) Heynh. lacking a dominant K+ transport system. J. Exp. Bot. 2001, 52, 839–844. [Google Scholar] [CrossRef] [Green Version]
- Sentenac, H.; Bonneaud, N.; Minet, M.; Lacroute, F.; Salmon, J.M.; Gaymard, F.; Grignon, C. Cloning and expression in yeast of a plant potassium ion transport system. Science 1992, 256, 663–665. [Google Scholar] [CrossRef] [PubMed]
- Alemán, F.; Nieves-Cordones, M.; Martínez, V.; Rubio, F. Root K+ Acquisition in Plants: The Arabidopsis thaliana Model. Plant Cell Physiol. 2011, 52, 1603–1612. [Google Scholar] [CrossRef]
- Lagarde, D.; Basset, M.; Lepetit, M.; Conejero, G.; Gaymard, F.; Astruc, S.; Grignon, C. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 1996, 9, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Pilot, G.; Gaymard, F.; Mouline, K.; Chérel, I.; Sentena, H. Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 2003, 5, 773–787. [Google Scholar] [CrossRef]
- Buschmann, P.H.; Vaidyanathan, R.; Gassmann, W.; Schroeder, J.I. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000, 122, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Cui, K.; Duan, A.; Zeng, Y.; Zhang, J. Genome-wide and molecular evolution analysis of the poplar KT/HAK/KUP potassium transporter gene family. Ecol. Evol. 2012, 2, 1996–2004. [Google Scholar] [CrossRef]
- Amrutha, R.N.; Sekhar, P.N.; Varshney, R.K.; Kishor, P.B.K. Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 2007, 172, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Hyun, T.K.; Rim, Y.; Kim, E.; Kim, J.S. Genome-wide andmolecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genom. 2014, 36, 365–374. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Lu, J.H.; Chen, D.; Zhang, J.; Qi, K.J.; Cheng, R.; Zhang, H.P.; Zhang, S.L. Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear. Genome 2018, 61, 755–765. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Li, M.; Oh, S.; Kronzucker, H.J. Capacity and plasticity of potassium channels and high-affinity transporters in roots of barley and Arabidopsis. Plant Physiol. 2013, 162, 496–511. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.C.; Harberd, N.P. An Arabidopsis Soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 2013, 25, 3535–3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyo, Y.J.; Gierth, M.; Cho, S.M.H. High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and post germination growth under low-potassium conditions. Plant Physiol. 2010, 153, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schachtman, D.P.; Schroeder, J.I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 1994, 370, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Rubio, F.L.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef]
- Horie, T.; Hauser, F.; Schroede, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, N.A.; Rubio, F. High-affinity potassium and sodium transport system in plants. J. Exp. Bot. 2006, 57, 1149–1160. [Google Scholar] [CrossRef] [Green Version]
- Garciadeblás, B.; Senn, M.E.; Baňuelo, M.A.; Rondríguez-Navarro, A. Sodium transport and HKT transporters: The rice model. Plant J. 2003, 34, 788–801. [Google Scholar] [CrossRef]
- Oomen, R.J.F.J.; Benito, B.; Sentenac, H.; Rodríguez-Navarro, A.; Talón, M.; Véry, A.A.; Domingo, C. HKT2;2/1, a K+-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 2012, 71, 750–762. [Google Scholar] [CrossRef] [Green Version]
- Almeida, P.; Katschnig, D.; Boer, A.H. HKT transporters-state of the art. Int. J. Mol. Sci. 2013, 14, 20359–20385. [Google Scholar] [CrossRef] [Green Version]
- Santa-Maria, G.E.; Rubio, F.; Dubcovsky, J.; Rodriguez-Navarro, A. The HAK1 Gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 1997, 9, 2281–2289. [Google Scholar]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wei, J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 2012, 39, 8465–8473. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cordero, M.A.; Martínez, V.; Rubio, F. Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol. Biol. 2004, 56, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Kispal, G.; Csere, P.; Prohl, C.; Lill, R. The mitochondrial proteins atm1p and nfs1p are essential for biogenesis of cytosolic FE/S proteins. EMBO (Eur. Mol. Biol. Organ.) J. 1999, 18, 3981–3989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beek, J.T.; Guskov, A.; Slotboom, D.J. Structural diversity of ABC transporters. J. Cell Biol. 2014, 143, 419–435. [Google Scholar]
- Lee, J.Y.; Yang, J.G.; Zhitnitsky, D.; Lewinson, O.; Rees, D.C. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 2014, 343, 1133–1135. [Google Scholar] [CrossRef] [Green Version]
- Yazaki, K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006, 580, 1183–1191. [Google Scholar] [CrossRef] [Green Version]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Song, W.Y.; Park, J.; Mendoza-Cózatl, D.G.; Suter-Grotemeyer, M.; Martinoia, E. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef] [Green Version]
- Sousa, C.A.; Hanselaer, S.; Soares, E.V. ABCC subfamily vacuolar transporters are involved in Pb (Lead) detoxification in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2015, 175, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.S.; Lai, R.; Jennings, J.L.; Link, A.J.; Hinnebusch, A.G. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol. Cell. Biol. 2005, 25, 9859–9873. [Google Scholar] [CrossRef] [Green Version]
- Jasinski, M.; Banasiak, J.; Radom, M.; Kalitkiewicz, A.; Figlerowicz, M. Full-Size ABC Transporters from the ABCG Subfamily in Medicago truncatula. Mol. Plant Microbe Interact. 2009, 22, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marr, I.L.; Suryana, N.; Lukulay, P.; Marr, M.I. Determination of chlorophyll a and b by simultaneous multi-component spectrophotometry. Fresenius J. Anal. Chem. 1995, 352, 456–460. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 40–425. [Google Scholar]
- Lewis, P.O.; Kumar, S.; Tamura, K.; Nei, M.; Lewis, P.O. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar]

| RawData | TotalReads | TotalBases | Error% | Q20% | Q30% | GC% |
|---|---|---|---|---|---|---|
| TP01-1 | 34,165,110 | 4,881,892,216 | 0.0134 | 97.5 | 91.74 | 51.45 |
| TP01-2 | 33,169,330 | 4,878,900,814 | 0.0138 | 97.3 | 91.73 | 51.40 |
| TP01-3 | 35,157,926 | 4,881,996,121 | 0.0136 | 97.4 | 91.78 | 51.47 |
| TP02-1 | 75,835,214 | 10,521,500,724 | 0.0103 | 98.7 | 95.40 | 47.4 |
| TP02-2 | 75,831,068 | 10,516,553,023 | 0.0101 | 98.2 | 95.27 | 46.9 |
| TP02-3 | 75,834,548 | 10,520,436,458 | 0.0102 | 98.6 | 95.35 | 47.3 |
| Parameter | Value | Parameter | Value |
|---|---|---|---|
| Radio frequency power | 1.45 V | Orifice of sampling cone | 1.0 mm |
| Carrier gas flow rate | 0.85 L/min | Orifice of skimmer cone | 0.4 mm |
| Mixed gas flow rate | 0.28 L/min | Sample depth | 7.0 mm |
| Plasma gas flow rate | 15.0 L/min | Sample uptake flow rate | 0.1 r/s |
| Auxiliary gas flow rate | 1.0 L/min | Sample uptake amount | 0.4 mL/min |
| Helium flow rate | 5 mL/min | Spray chamber temperature | 2 °C |
| Seq_ID | Gene Name | Primer(5’-3’) |
|---|---|---|
| c126889g1_i1 | PaHAK5 | P1 ATGGATCTCAGCAGGATGGG |
| P2 TTCCATTGTCGTTTGCCCTG | ||
| c219737g1_i1 | PaKT2 | P1 GAGCCTTCTGCCAAAC |
| P2 CCATCTCCAACCACCA | ||
| c56511g1_i1 | PaKT4 | P1 GGTCGGTGTACGAGGAT |
| P2 TGAGGCTAATGTGAGGAA | ||
| c123766g1_i1 | PaKUP6 | P1 CGGGACCCTCAAGAAG |
| P2 GCCTATGCCACGGACT | ||
| c125123g6_i1 | PaKUP7 | P1 TCTGAGGCTATGTTTGCCGA |
| P2 GTAGCTGTCGTCATTGCTCG | ||
| c108651g2_i1 | PaAKT1 | P1 TTCAGAAGGGTTGCAACAGC |
| P2 AAATTCCACGGCTCCAGAGA | ||
| c121941g1_i1 | PaAKT2 | P1 GGAGCAGGGAAATGTTGTGG |
| P2 TAAGGCTGAGGGATGTTGCA | ||
| c72107g1_i1 | PaHKT1 | P1 CGGTTCTTGTGCTGTT |
| P2 TGCTTCTTGTCGCTGA | ||
| Actin | P1 CTTGACTTTGAGCAGGAATCGGAGA | |
| P2 ACCTGCTGCTTCCATACCTATCAAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Q.; Ma, L.; Tan, P.; Deng, W.; Huang, C.; Liu, D.; Lin, W.; Su, Y. Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca acinosa Roxb. Plants 2020, 9, 470. https://doi.org/10.3390/plants9040470
Xie Q, Ma L, Tan P, Deng W, Huang C, Liu D, Lin W, Su Y. Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca acinosa Roxb. Plants. 2020; 9(4):470. https://doi.org/10.3390/plants9040470
Chicago/Turabian StyleXie, Qin, Liying Ma, Peng Tan, Wentao Deng, Chao Huang, Deming Liu, Wanhuang Lin, and Yi Su. 2020. "Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca acinosa Roxb" Plants 9, no. 4: 470. https://doi.org/10.3390/plants9040470





