Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement
Abstract
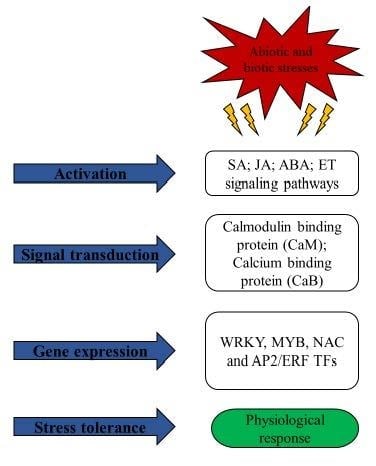
1. Introduction
2. WRKY TFs in Response to Stresses
2.1. Classification and Diversification of WRKY Genes Family
2.2. Function and Expression Pattern of WRKYs under Abiotic Stresses
2.3. Function and Expression Pattern of WRKY Genes under Biotic Stresses
3. NAC TFs in Response to Stresses
3.1. Classification and Diversification of NAC Gene Family
3.2. Function and Expression Pattern of NAC TFs under Abiotic Stresses
3.3. Function and Expression Pattern of NAC TFs under Biotic Stresses
4. MYB TFs in Response to Stresses
4.1. Classification and Diversification of MYB Gene Family
4.2. Function and Expression Pattern of MYB TFs under Abiotic Stresses
4.3. Function and Expression Pattern of MYB TFs under Biotic Stresses
5. AP2/ERF TFs in Response to Stresses
5.1. Classification and Diversification of AP2/ERF Gene Family
5.2. Function and Expression Pattern of AP2/ERF TFs under Abiotic Stresses
5.3. Function and Expression Pattern of AP2/ERF TFs under Biotic Stresses
6. Conclusion and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Bhadouria, R.; Singh, R.; Singh, V.K.; Borthakur, A.; Ahamad, A.; Kumar, G.; Singh, P. Agriculture in the era of climate change: Consequences and effects. In Climate Change and Agricultural Ecosystems; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–23. [Google Scholar]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Khan, M.; Sharif, R.; Mujtaba, M.; Gao, S.-J. Sugarcane Omics: An update on the current status of research and crop improvement. Plants 2019, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Fraire-Velázquez, S.; Rodríguez-Guerra, R.; Sánchez-Calderón, L. Abiotic and biotic stress response crosstalk in plants. In Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives; IntechOpen: London, UK, 2011; pp. 3–26. [Google Scholar]
- Gonzalez, D.H. Introduction to transcription factor structure and function. In Plant Transcription Factors; Elsevier: Amsterdam, The Netherlands, 2016; pp. 3–11. [Google Scholar]
- Inukai, S.; Kock, K.H.; Bulyk, M.L. Transcription factor–DNA binding: Beyond binding site motifs. Curr. Opin. Genet. Deve. 2017, 43, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant. Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Mustafa, G.; Joyia, F.A.; Anwar, S.; Parvaiz, A.; Khan, M.S. Biotechnological interventions for the improvement of sugarcane crop and sugar production. In Sugarcane-Technology and Research; IntechOpen: London, UK, 2018; pp. 113–138. [Google Scholar]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant. Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant. Biol. 2015, 15, 66. [Google Scholar] [CrossRef]
- Bao, W.; Wang, X.; Chen, M.; Chai, T.; Wang, H. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant. Cell Rep. 2018, 37, 1033–1048. [Google Scholar] [CrossRef]
- Li, Z.; Hua, X.; Zhong, W.; Yuan, Y.; Wang, Y.; Wang, Z.; Ming, R.; Zhang, J. Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant. Cell Physiol. 2020, 61, 616–630. [Google Scholar] [CrossRef]
- Uluhan, E.; Keleş, E.N.; Tufan, F. Analysis of WRKY transcription factors in barley cultivars infected with Fusarium culmorum. Int. J. Life Sci. Biotechnol. 2019, 2, 165–174. [Google Scholar] [CrossRef]
- Lilly, J.J.; Subramanian, B. Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa L.). Plant. Sci. 2019, 280, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Xu, X.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.). Gene 2013, 524, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Yang, J.-F.; Li, M.; Xu, Z.-S.; Fu, J.-D. The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, J.; Shang, X.; Geng, M.; Gao, J.; Zhao, S.; Yu, X.; Liu, D.; Kang, Z.; Wang, X. WRKY transcription factors shared by BTH-induced resistance and NPR1-mediated acquired resistance improve broad-spectrum disease resistance in wheat. Mol. Plant. Microbe Interact. 2019, 33, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Wei, H.; Wang, H.; Su, J.; Yu, S. Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genet. 2018, 19, 48. [Google Scholar] [CrossRef]
- Goyal, P.; Manzoor, M.M.; Vishwakarma, R.A.; Sharma, D.; Dhar, M.K.; Gupta, S. A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant. Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant. Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- He, G.-H.; Xu, J.-Y.; Wang, Y.-X.; Liu, J.-M.; Li, P.-S.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant. Biol. 2016, 16, 116. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant. Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Mishra, V.K.; Kumari, S.; Chand, R.; Varadwaj, P.K. Deciphering genome-wide WRKY gene family of Triticum aestivum L. and their functional role in response to abiotic stress. Genes Genom. 2019, 41, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, T.; Sun, X.; Wang, Y.; Du, C.; Zhu, Z.; Gichuki, D.K.; Wang, Q.; Li, S.; Xin, H. Overexpression of VaWRKY12, a transcription factor from Vitis amurensis with increased nuclear localization under low temperature, enhances cold tolerance of plants. Plant. Mol. Biol. 2019, 100, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Yadav, G.; Jat, H. Integrated farming system approaches for managing saline and waterlogged ecologies. In Research Developments in Saline Agriculture; Springer: Berlin/Heidelberg, Germany, 2019; pp. 753–768. [Google Scholar]
- Hsu, F.-C.; Chou, M.-Y.; Chou, S.-J.; Li, Y.-R.; Peng, H.-P.; Shih, M.-C. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant. Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef]
- Raineri, J.; Ribichich, K.F.; Chan, R.L. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant. Cell Rep. 2015, 34, 2065–2080. [Google Scholar] [CrossRef]
- Han, C.; Li, J.; Ma, Y.; Guo, J.; Guo, X.; Xu, J. PlWRKY70: A Paeonia lactiflora transcription factor that sensitively responds to low-temperature, salt and waterlogging stresses. Can. J. Plant. Sci. 2019, 100, 146–155. [Google Scholar] [CrossRef]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.-J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef]
- Babitha, K.; Ramu, S.; Pruthvi, V.; Mahesh, P.; Nataraja, K.N.; Udayakumar, M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013, 22, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant. Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, F.; Zhang, X.; Wang, W.; Sun, T.; Chen, Y.; Dai, M.; Yu, S.; Xu, L.; Su, Y.; et al. Expression characteristics and functional analysis of the ScWRKY3 gene from sugarcane. Int. J. Mol. Sci. 2018, 19, 4059. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Tian, Y.; Liu, X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant. Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Gu, L.; Ma, Q.; Zhang, C.; Wang, C.; Wei, H.; Wang, H.; Yu, S. The Cotton GhWRKY91 Transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front. Plant. Sci. 2019, 10, 1352. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.W.; Bian, X.H.; Shen, M.; Xiao, J.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant. J. 2019, 100, 384–398. [Google Scholar] [CrossRef]
- Thomas, T.D.; Dinakar, C.; Puthur, J.T. Effect of UV-B priming on the abiotic stress tolerance of stress-sensitive rice seedlings: Priming imprints and cross-tolerance. Plant. Physiol. Biochem. 2020, 147, 21–30. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant. Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Marchica, A.; Lorenzini, G.; Papini, R.; Bernardi, R.; Nali, C.; Pellegrini, E. Signalling molecules responsive to ozone-induced oxidative stress in Salvia officinalis. Sci. Total Environ. 2019, 657, 568–576. [Google Scholar] [CrossRef]
- Garg, N.; Manchanda, G. ROS generation in plants: Boon or bane? Plant. Biosys. 2009, 143, 81–96. [Google Scholar] [CrossRef]
- Luan, Q.; Chen, C.; Liu, M.; Li, Q.; Wang, L.; Ren, Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant. Sci. 2019, 279, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Léon, C.; Kappel, C.; Coutos-Thévenot, P.; Corio-Costet, M.-F.; Delrot, S.; Lauvergeat, V. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiologia Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Wang, N.; Guo, X.; Gao, Z. GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant. Cell. Tiss. Org. Cult. 2015, 121, 127–140. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef]
- Jing, Z.; Liu, Z. Genome-wide identification of WRKY transcription factors in kiwifruit (Actinidia spp.) and analysis of WRKY expression in responses to biotic and abiotic stresses. Genes Genom. 2018, 40, 429–446. [Google Scholar] [CrossRef]
- Dong, H.; Tan, J.; Li, M.; Yu, Y.; Jia, S.; Zhang, C.; Wu, Y.; Liu, Y. Transcriptome analysis of soybean WRKY TFs in response to Peronospora manshurica infection. Genomics 2019, 111, 1412–1422. [Google Scholar] [CrossRef]
- Ntambo, M.S.; Meng, J.-Y.; Rott, P.C.; Henry, R.J.; Zhang, H.-L.; Gao, S.-J. Comparative transcriptome profiling of resistant and susceptible sugarcane cultivars in response to infection by Xanthomonas albilineans. Int. J. Mol. Sci. 2019, 20, 6138. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef]
- Sablowski, R.W.; Meyerowitz, E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 1998, 92, 93–103. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant. Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant. Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant. Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Skriver, K.; Larsen, S.; Leggio, L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, S.-G.; Kim, Y.-S.; Seo, P.J.; Bae, M.; Yoon, H.-K.; Park, C.-M. Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 2007, 35, 203–213. [Google Scholar] [CrossRef]
- Liang, M.; Li, H.; Zhou, F.; Li, H.; Liu, J.; Hao, Y.; Wang, Y.; Zhao, H.; Han, S. Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 2015, 16, 1062–1074. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Bita, C.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant. Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Ditt, R.F.; Gentile, A.; Tavares, R.G.; Camargo, S.R.; Fernandez, J.H.; Silva, M.J.d.; Menossi, M. Analysis of the stress-inducible transcription factor SsNAC23 in sugarcane plants. Sci. Agric. 2011, 68, 454–461. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant. J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant. Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; Zhang, Y.; Lyu, Y. A Stress-Responsive NAC transcription factor from tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 3225. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-m.; Zhang, H.-f.; Liu, S.-y.; Wang, X.-k.; Zhang, Y.-m.; Meng, Y.-c.; Luo, D.; Chen, R.-g. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant. Sci. 2020, 291, 110346. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Shukla, A. Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 2015, 22, 147–161. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant. Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.-H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.-K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant. Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice Os NAC 6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant. Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Rahman, H.; Ramanathan, V.; Nallathambi, J.; Duraialagaraja, S.; Muthurajan, R. Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant. Sci. 2016, 6, 1174. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant. Biol. 2019, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Gunapati, S.; Naresh, R.; Ranjan, S.; Nigam, D.; Hans, A.; Verma, P.C.; Gadre, R.; Pathre, U.V.; Sane, A.P.; Sane, V.A. Expression of GhNAC2 from G. herbaceum, improves root growth and imparts tolerance to drought in transgenic cotton and Arabidopsis. Sci. Rep. 2016, 6, 24978. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Li, K.-Q.; Xu, X.-Y.; Zhang, H.-P.; Chen, H.-X.; Chen, Y.-H.; Hao, J.; Wang, Y.; Huang, X.-S.; Zhang, S.-L. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant. Sci. 2017, 8, 1049. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, Y.-S.; Han, S.-H.; Lee, B.-D.; Paek, N.-C. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant. Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, Y.; Li, H.; Wang, T.; Zhang, J.; Ouyang, B.; Ye, Z. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites. Plant. Sci. 2018, 271, 9–19. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant. J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 2014, 9, 1–15. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant. Biol. 2015, 15, 268. [Google Scholar] [CrossRef]
- He, Z.; Li, Z.; Lu, H.; Huo, L.; Wang, Z.; Wang, Y.; Ji, X. The NAC protein from Tamarix hispida, ThNAC7, confers salt and osmotic stress tolerance by increasing reactive oxygen species scavenging capability. Plants 2019, 8, 221. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopath. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Sun, Y.-F.; Zhu, L.; Xu, L.-S.; Chen, X.-M.; Liu, B.; Yu, Y.-T.; Wang, X.-J.; Huang, L.-L. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant. Pathol. 2010, 74, 394–402. [Google Scholar] [CrossRef]
- Voitsik, A.-M.; Muench, S.; Deising, H.B.; Voll, L.M. Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant. Biol. 2013, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant. Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-M.; Zhang, Q.; Pei, C.-L.; Li, X.; Huang, X.-L.; Chang, C.-Y.; Wang, X.-J.; Huang, L.-L.; Kang, Z.-S. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiol. Mol. Plant. P. 2018, 102, 144–153. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant. Physiol. Biochem. 2017, 120, 61–74. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant. Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant. Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef]
- Meisrimler, C.N.; Pelgrom, A.J.; Oud, B.; Out, S.; Van den Ackerveken, G. Multiple downy mildew effectors target the stress-related NAC transcription factor LsNAC069 in lettuce. Plant. J. 2019, 99, 1098–1115. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant. Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Hajiebrahimi, A.; Owji, H.; Hemmati, S. Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 2017, 60, 797–814. [Google Scholar] [CrossRef]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178. [Google Scholar] [CrossRef]
- Mmadi, M.A.; Dossa, K.; Wang, L.; Zhou, R.; Wang, Y.; Cisse, N.; Sy, M.O.; Zhang, X. Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 2017, 8, 362. [Google Scholar] [CrossRef]
- Meng, X.; Wang, J.-R.; Wang, G.-D.; Liang, X.-Q.; Li, X.-D.; Meng, Q.-W. An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J. Plant. Physiol. 2015, 175, 1–8. [Google Scholar] [CrossRef]
- Deeba, F.; Sultana, T.; Javaid, B.; Mahmood, T.; Naqvi, S. Molecular characterization of a MYB protein from Oryza sativa for its role in abiotic stress tolerance. Braz. Arch. Biol. Technol. 2017, 60, 1–12. [Google Scholar] [CrossRef]
- Casaretto, J.A.; El-kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.-M.; Rothstein, S.J. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genom. 2016, 17, 312. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, X.; Wang, F.; Zhang, L.; Xin, M.; Hu, Z.; Yao, Y.; Ni, Z.; Sun, Q.; Peng, H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant. Biol. 2017, 17, 208. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Liu, Y.; Zhao, L.; Zhang, S.; Huang, X. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant. Cell Environ. 2019, 42, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Seo, J.S.; Sohn, H.B.; Noh, K.; Jung, C.; An, J.H.; Donovan, C.M.; Somers, D.A.; Kim, D.I.; Jeong, S.-C.; Kim, C.-G. Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol. Breed. 2012, 29, 601–608. [Google Scholar] [CrossRef]
- Chen, B.-J.; Wang, Y.; Hu, Y.-L.; Wu, Q.; Lin, Z.-P. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant. Sci. 2005, 168, 493–500. [Google Scholar] [CrossRef]
- Yu, Y.-T.; Wu, Z.; Lu, K.; Bi, C.; Liang, S.; Wang, X.-F.; Zhang, D.-P. Overexpression of the MYB37 transcription factor enhances abscisic acid sensitivity, and improves both drought tolerance and seed productivity in Arabidopsis thaliana. Plant. Mol. Biol. 2016, 90, 267–279. [Google Scholar] [CrossRef]
- Prabu, G.; Prasad, D.T. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant. Cell Rep. 2012, 31, 661–669. [Google Scholar] [CrossRef]
- Fávero, P.J.R.; Mara, D.A.L.; Dos, S.B.M. Overexpression of ScMYBAS1 alternative splicing transcripts differentially impacts biomass accumulation and drought tolerance in rice transgenic plants. PLoS ONE 2018, 13, e0207534. [Google Scholar]
- Guo, J.; Ling, H.; Ma, J.; Chen, Y.; Su, Y.; Lin, Q.; Gao, S.; Wang, H.; Que, Y.; Xu, L. A sugarcane R2R3-MYB transcriprion factor gene is alternatively spliced during drought stress. Sci. Rep. 2017, 7, 41922. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.-H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant. Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant. 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Li, X.W.; Wang, Y.; Yan, F.; Li, J.W.; Zhao, Y.; Zhao, X.; Zhai, Y.; Wang, Q.Y. Overexpression of soybean R2R3-MYB transcription factor, GmMYB12B2, and tolerance to UV radiation and salt stress in transgenic Arabidopsis. Genet. Mol. Res. 2016, 15, 6573. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, J.; Ma, Z.; Wang, J.; Zhou, C. Arabidopsis transcription factor MYB102 increases plant susceptibility to aphids by substantial activation of ethylene biosynthesis. Biomolecules 2018, 8, 39. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef]
- Segarra, G.; Van der Ent, S.; Trillas, I.; Pieterse, C. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant. Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Ibraheem, F.; Gaffoor, I.; Tan, Q.; Shyu, C.-R.; Chopra, S. A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize. Molecules 2015, 20, 2388–2404. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Sheng, L.; Du, X.; Wang, Y.; Zhang, Y.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic. Res. 2019, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Zhang, C.-L.; Wang, G.-L.; Wang, Y.-X.; Qi, C.-H.; Zhao, Q.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant. Biol. 2019, 19, 362. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Hussain, A.; Adnan, M.; Khan, M.I.; Ashraf, M.F.; Zainab, M.; Khan, K.A.; Ghramh, H.A.; He, S. A novel MYB transcription factor CaPHL8 provide clues about evolution of pepper immunity againstsoil borne pathogen. Microb. Pathogen. 2019, 137, 103758. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, D.; Li, G.; Yang, Y.; Zhang, G.; Li, S.; Liang, Z. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2). BMC Plant. Biol. 2019, 19, 478. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740. [Google Scholar] [CrossRef]
- Dong, C.-J.; Liu, J.-Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant. Biol. 2010, 10, 47. [Google Scholar] [CrossRef]
- Xiong, A.-S.; Jiang, H.-H.; Zhuang, J.; Peng, R.-H.; Jin, X.-F.; Zhu, B.; Wang, F.; Zhang, J.; Yao, Q.-H. Expression and function of a modified AP2/ERF transcription factor from Brassica napus enhances cold tolerance in transgenic Arabidopsis. Mol. Biotechnol. 2013, 53, 198–206. [Google Scholar] [CrossRef]
- Zhao, T.-J.; Sun, S.; Liu, Y.; Liu, J.-M.; Liu, Q.; Yan, Y.-B.; Zhou, H.-M. Regulating the drought-responsive element (DRE)-mediated signaling pathway by synergic functions of trans-active and trans-inactive DRE binding factors in Brassica napus. J. Biol. Chem. 2006, 281, 10752–10759. [Google Scholar] [CrossRef] [PubMed]
- Illgen, S.; Zintl, S.; Zuther, E.; Hincha, D.; Schmulling, T. Characterisation of the ERF102 to ERF105 genes of Arabidopsis thaliana and their role in the response to cold stress. Plant Mol. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, Z.; Zhang, L.; Fang, L.; Zhang, J.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. Overexpression of ethylene response factors VaERF080 and VaERF087 from Vitis amurensis enhances cold tolerance in Arabidopsis. Sci. Hortic. 2019, 243, 320–326. [Google Scholar] [CrossRef]
- Lv, K.; Li, J.; Zhao, K.; Chen, S.; Nie, J.; Zhang, W.; Liu, G.; Wei, H. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant. Sci. 2020, 292, 110375. [Google Scholar] [CrossRef]
- Moon, S.-J.; Min, M.K.; Kim, J.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.-G. Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front. Plant. Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Gomathi, R.; Rao, P.N.G.; Chandran, K.; Selvi, A. Adaptive responses of sugarcane to waterlogging stress: An over view. Sugar Tech 2015, 17, 325–338. [Google Scholar] [CrossRef]
- Du, H.; Huang, M.; Zhang, Z.; Cheng, S. Genome-wide analysis of the AP2/ERF gene family in maize waterlogging stress response. Euphytica 2014, 198, 115–126. [Google Scholar] [CrossRef]
- Shahzad, Z.; Canut, M.; Tournaire-Roux, C.; Martiniere, A.; Boursiac, Y.; Loudet, O.; Maurel, C. A potassium-dependent oxygen sensing pathway regulates plant root hydraulics. Cell 2016, 167, 87–98. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant. Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant. Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Fan, W.; Yang, Y.; Wang, Z.; Yin, Y.; Yu, C.; Shi, Q.; Guo, J.; Xuan, L.; Hua, J. Molecular cloning and expression analysis of three ThERFs involved in the response to waterlogging stress of Taxodium ‘Zhongshanshan406’, and subcellular localization of the gene products. PeerJ 2018, 6, e4434. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.-L.; Ma, J.-T.; Zhao, Y.-M.; Wei, Y.-H.; Tang, Y.-X.; Wu, Y.-M. Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. Gene 2012, 506, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.-Y.; Cheng, X.-G.; Xu, Z.-S.; Li, L.-C.; Ye, X.-G.; Xia, L.-Q.; Ma, Y.-Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, S.; Watanabe, K.; Ohori, T.; Moriwaki, T.; Maruyama, K.; Mizoi, J.; Myint Phyu Sin Htwe, N.; Fujita, Y.; Sekita, S.; Shinozaki, K. Soybean DREB 1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant. J. 2015, 81, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant. Biol. 2016, 16, 171. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Y.; Wang, W.; Zhao, X.; Qin, Q.; Sun, F.; Hu, F.; Zhao, Y.; Li, Z.; Fu, B. Characterization of transcription factor gene OsDRAP1 conferring drought tolerance in rice. Front. Plant. Sci. 2018, 9, 94. [Google Scholar] [CrossRef]
- de Souza, W.R.; de Oliveira, N.G.; Vinecky, F.; Ribeiro, A.P.; Basso, M.F.; Casari, R.A.d.C.N.; da Cunha, B.A.D.B.; Duarte, K.E.; Santiago, T.R.; Martins, P.K. Field evaluation of AtDREB2A CA overexpressing sugarcane for drought tolerance. J. Agron. Crop. Sci. 2019, 205, 545–553. [Google Scholar] [CrossRef]
- Tavakol, E.; Sardaro, M.L.S.; Shariati, V.; Rossini, L.; Porceddu, E. Isolation, promoter analysis and expression profile of Dreb2 in response to drought stress in wheat ancestors. Gene 2014, 549, 24–32. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Xiao, H.-m.; Yang, Y. Characterization of TaDREB1 in wheat genotypes with different seed germination under osmotic stress. Hereditas 2018, 155, 26. [Google Scholar] [CrossRef]
- Yang, Y.; Al-Baidhani, H.H.J.; Harris, J.; Riboni, M.; Li, Y.; Mazonka, I.; Bazanova, N.; Chirkova, L.; Sarfraz Hussain, S.; Hrmova, M. DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: Impact on plant development, stress tolerance and yield. Plant. Biotechnol. J. 2020 18, 829–844. [CrossRef]
- Trujillo, L.E.; Sotolongo, M.; Menendez, C.; Ochogavia, M.E.; Coll, Y.; Hernandez, I.; Borras, H.O.; Thomma, B.P.H.J.; Vera, P.; Hernandez, L. SodERF3, a novel sugarcane ethylene responsive factor (ERF) enhances salt and drought tolerance when overexpressed in tobacco plants. Plant. Cell Physiol. 2008, 49, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, L.; Liu, X.; Cai, S.; Xu, H.; Huang, R.; Li, J.; Wei, X.; Zhang, Z. The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant. Physiol. 2014, 164, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithöfer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, B.-L.; Sun, S.; Xing, G.-M.; Wang, F.; Li, M.-Y.; Tian, Y.-S.; Xiong, A.-S. AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant. Genom. 2016, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chang, X.; Qi, D.; Dong, L.; Wang, G.; Fan, S.; Jiang, L.; Cheng, Q.; Chen, X.; Han, D. A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant. Sci. 2017, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae. Plant. Physiol. Biochem. 2019, 135, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Won, S.Y.; Suh, S.C.; Kim, H.; Wing, R.; Jeong, Y.; Hwang, I.; Kim, M. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 2007, 225, 575–588. [Google Scholar] [CrossRef]
- Dong, N.; Liu, X.; Lu, Y.; Du, L.; Xu, H.; Liu, H.; Xin, Z.; Zhang, Z. Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genomic. 2010, 10, 215–226. [Google Scholar] [CrossRef]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE 2013, 8, 1–12. [Google Scholar] [CrossRef]
- Lai, Y.; Dang, F.; Lin, J.; Yu, L.; Lin, J.; Lei, Y.; Chen, C.; Liu, Z.; Qiu, A.; Mou, S. Overexpression of a pepper CaERF5 gene in tobacco plants enhances resistance to Ralstonia solanacearum infection. Funct. Plant. Biol. 2014, 41, 758–767. [Google Scholar] [CrossRef]
- Wang, J.H.; Gu, K.D.; Han, P.L.; Yu, J.Q.; Wang, C.K.; Zhang, Q.Y.; You, C.X.; Hu, D.G.; Hao, Y.J. Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant. Sci. 2020, 291, 110351. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Bokowiec, M.T.; Laudeman, T.W.; Brannock, J.F.; Chen, X.; Timko, M.P. TOBFAC: The database of tobacco transcription factors. BMC Bioinform. 2008, 9, 53. [Google Scholar] [CrossRef] [PubMed]
| Plant Species | Type of Organism | WRKY | NAC | MYB | AP2/ERF |
|---|---|---|---|---|---|
| Saccharum spp. (Sugarcane) | Monocot plant | 39 | 44 | 36 | 6 |
| Sorghum bicolor (Sorghum) | Monocot plant | 134 | 180 | 145 | 42 |
| Triticum aestivum (Wheat) | Monocot plant | 171 | 263 | 263 | 43 |
| Oryza sativa subsp. japonica (Rice) | Monocot plant | 128 | 170 | 130 | 22 |
| Hordeum vulgare (Barley) | Monocot plant | 126 | 150 | 99 | 34 |
| Zea mays (Maize) | Monocot plant | 161 | 189 | 203 | 54 |
| Zoysia matrella (Manila grass) | Monocot plant | 269 | 313 | 293 | 49 |
| Brachypodium distachyon (False brome) | Monocot plant | 134 | 186 | 163 | 64 |
| Beta vulgaris (Sugar beet) | Dicot Plant | 49 | 59 | 68 | 14 |
| Arabidopsis thaliana | Dicot Plant | 90 | 138 | 168 | 30 |
| Nicotiana tabacum (Tobacco) | Dicot Plant | 210 | 280 | 319 | 93 |
| Jatropha curcas (Physic nut | Dicot Plant | 61 | 97 | 115 | 19 |
| Cucumis sativus (Cucumber) | Dicot Plant | 88 | 102 | 134 | 27 |
| Solanum lycopersicum (Tomato) | Dicot Plant | 81 | 101 | 140 | 27 |
| Vitis Vinifers (Grape) | Dicot Plant | 59 | 71 | 138 | 19 |
| Gossypium hirsutum (Cotton) | Dicot Plant | 238 | 306 | 441 | 59 |
| Glycine max (Soybean) | Dicot Plant | 296 | 269 | 430 | 99 |
| Sesamum indicum (Sesame) | Dicot Plant | 88 | 105 | 168 | 43 |
| Camelina sativa (Flax) | Dicot Plant | 224 | 350 | 402 | 93 |
| Brassica napus (Rapeseed) | Dicot Plant | 285 | 411 | 489 | 57 |
| Chlamydomonas reinhardtii | Single-celled green alga | 2 | 0 | 16 | 12 |
| Gonium pectoral | Single-celled green alga | 1 | 0 | 12 | 3 |
| Ostreococcus lucimarinus | Single-celled green alga | 2 | 0 | 11 | 3 |
| Selaginella moellendorffii | Moss | 19 | 22 | 24 | 10 |
| Physcomitrella patens | Moss | 117 | 142 | 180 | 44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants 2020, 9, 491. https://doi.org/10.3390/plants9040491
Javed T, Shabbir R, Ali A, Afzal I, Zaheer U, Gao S-J. Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants. 2020; 9(4):491. https://doi.org/10.3390/plants9040491
Chicago/Turabian StyleJaved, Talha, Rubab Shabbir, Ahmad Ali, Irfan Afzal, Uroosa Zaheer, and San-Ji Gao. 2020. "Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement" Plants 9, no. 4: 491. https://doi.org/10.3390/plants9040491
APA StyleJaved, T., Shabbir, R., Ali, A., Afzal, I., Zaheer, U., & Gao, S.-J. (2020). Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants, 9(4), 491. https://doi.org/10.3390/plants9040491