Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance
Abstract
1. Introduction
2. Results
2.1. Verification of Sequencing Data
2.2. Identification of Large Structural Variants
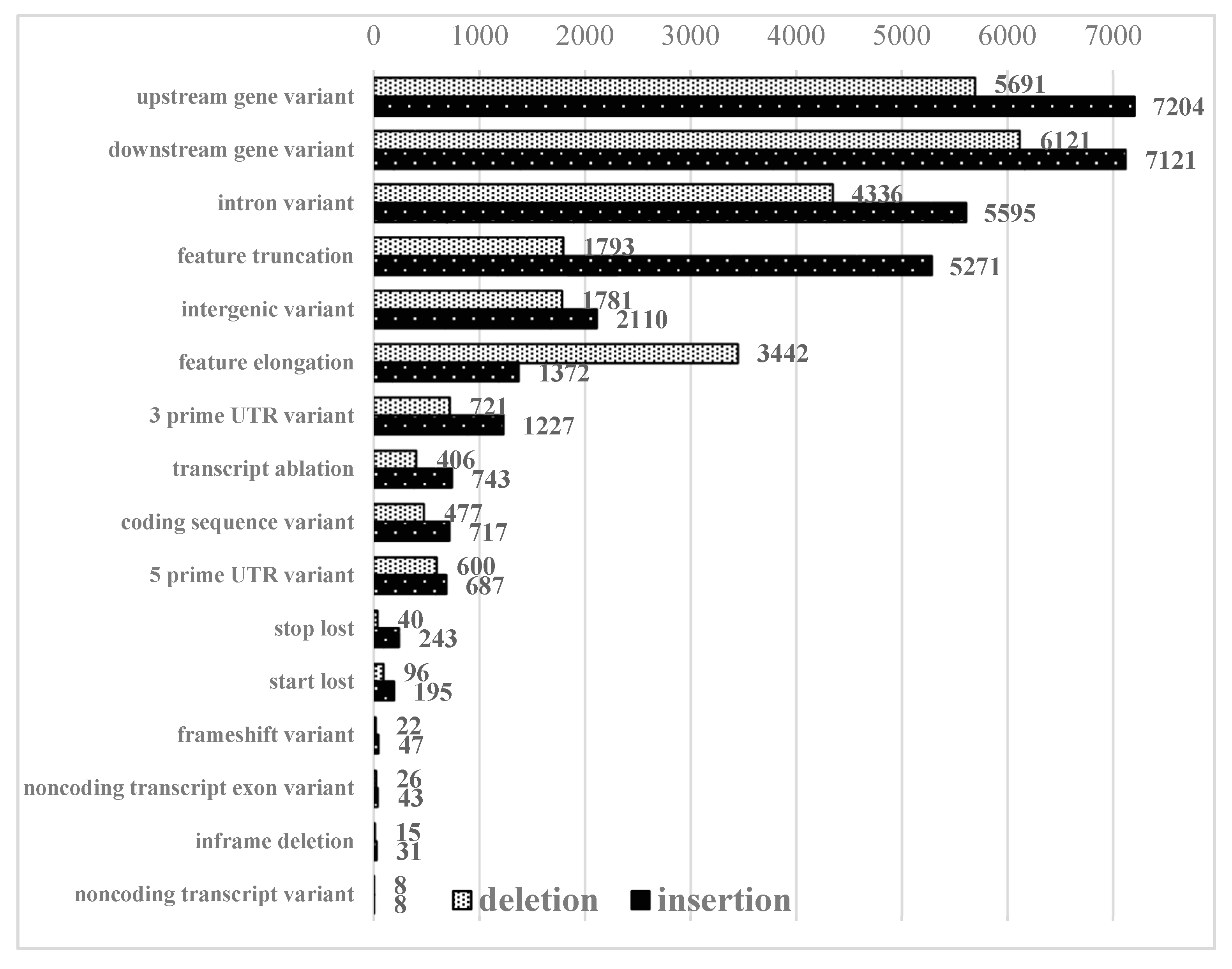
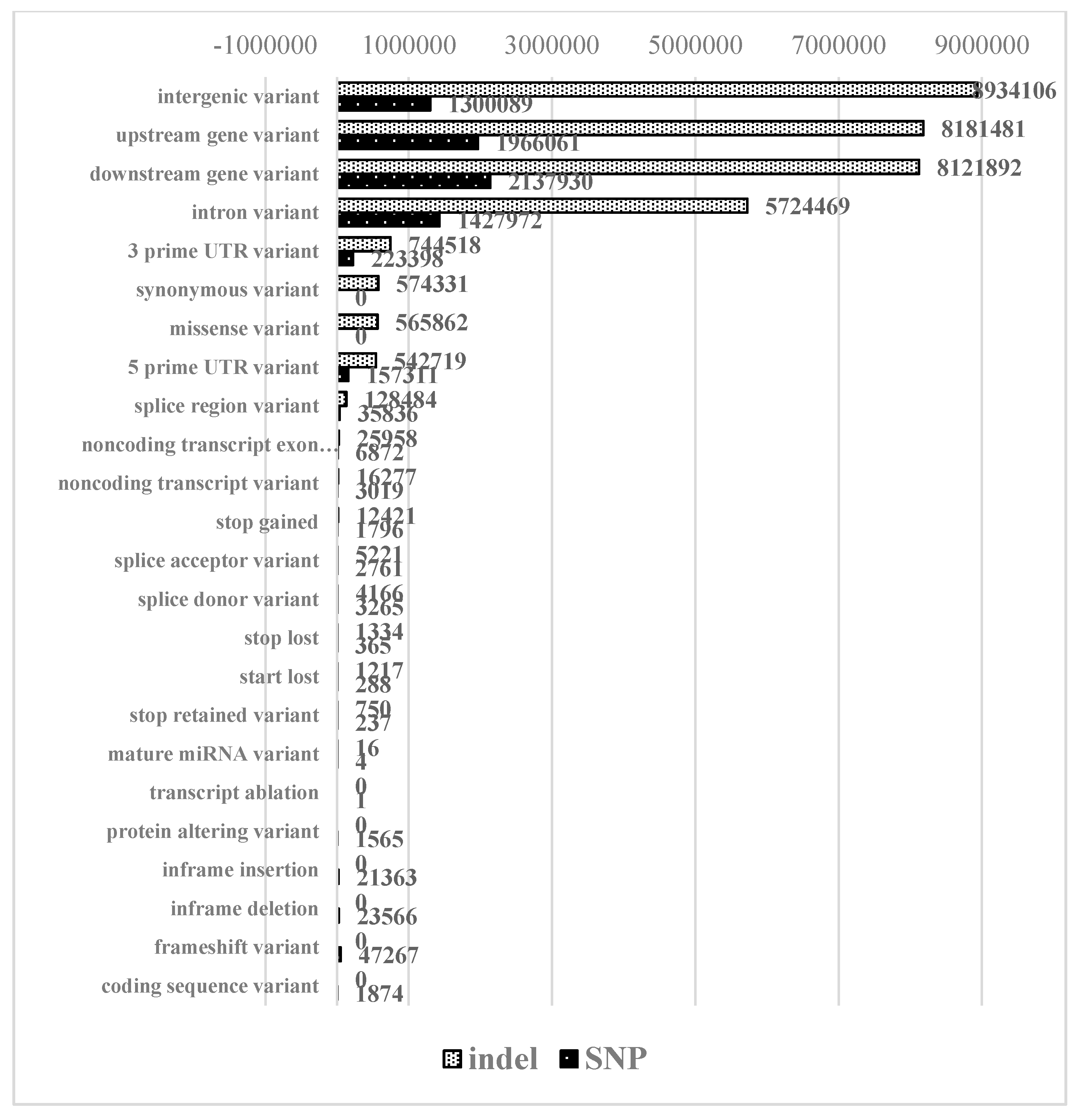
2.3. Identification of Small Structural Changes
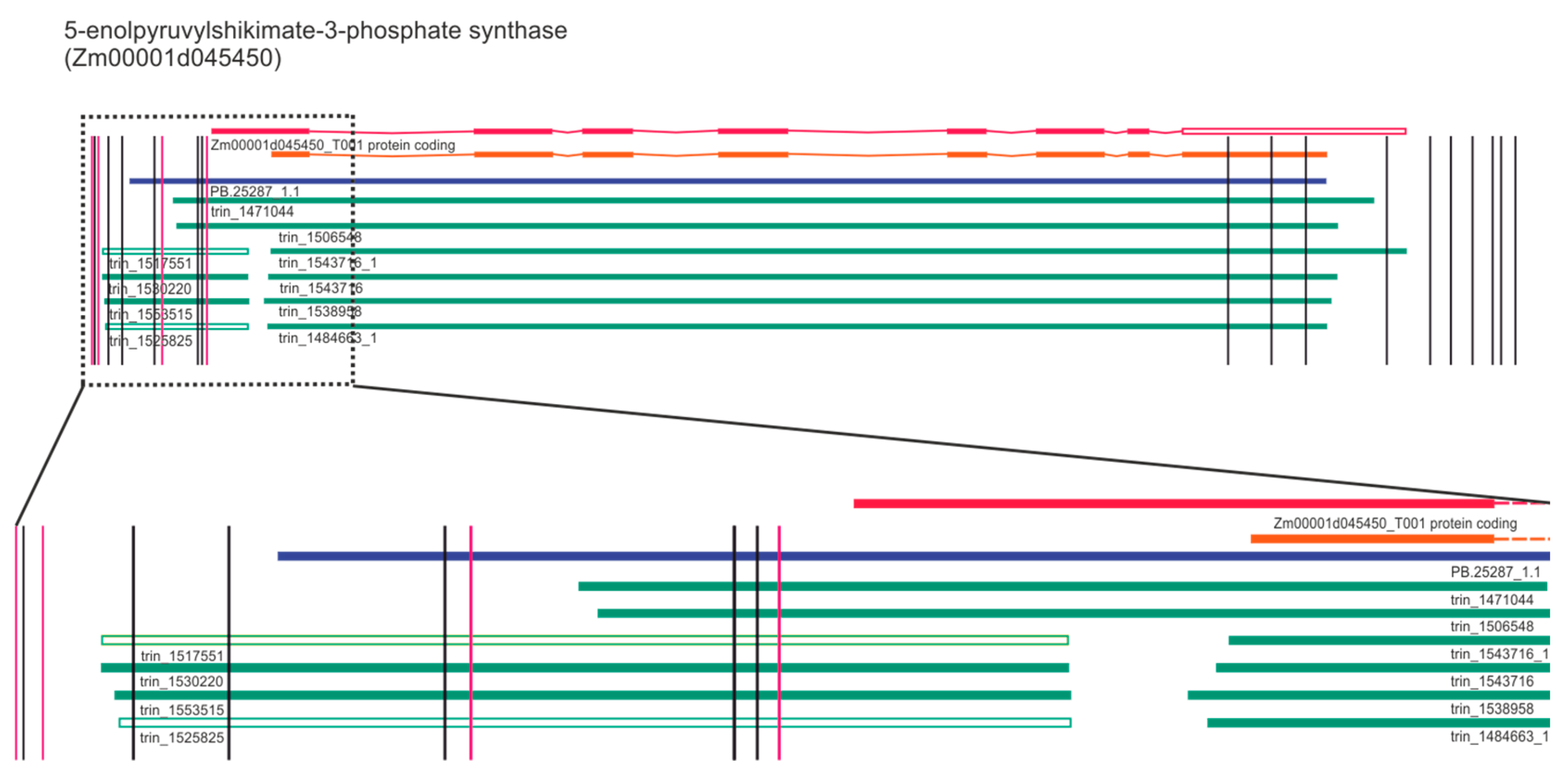
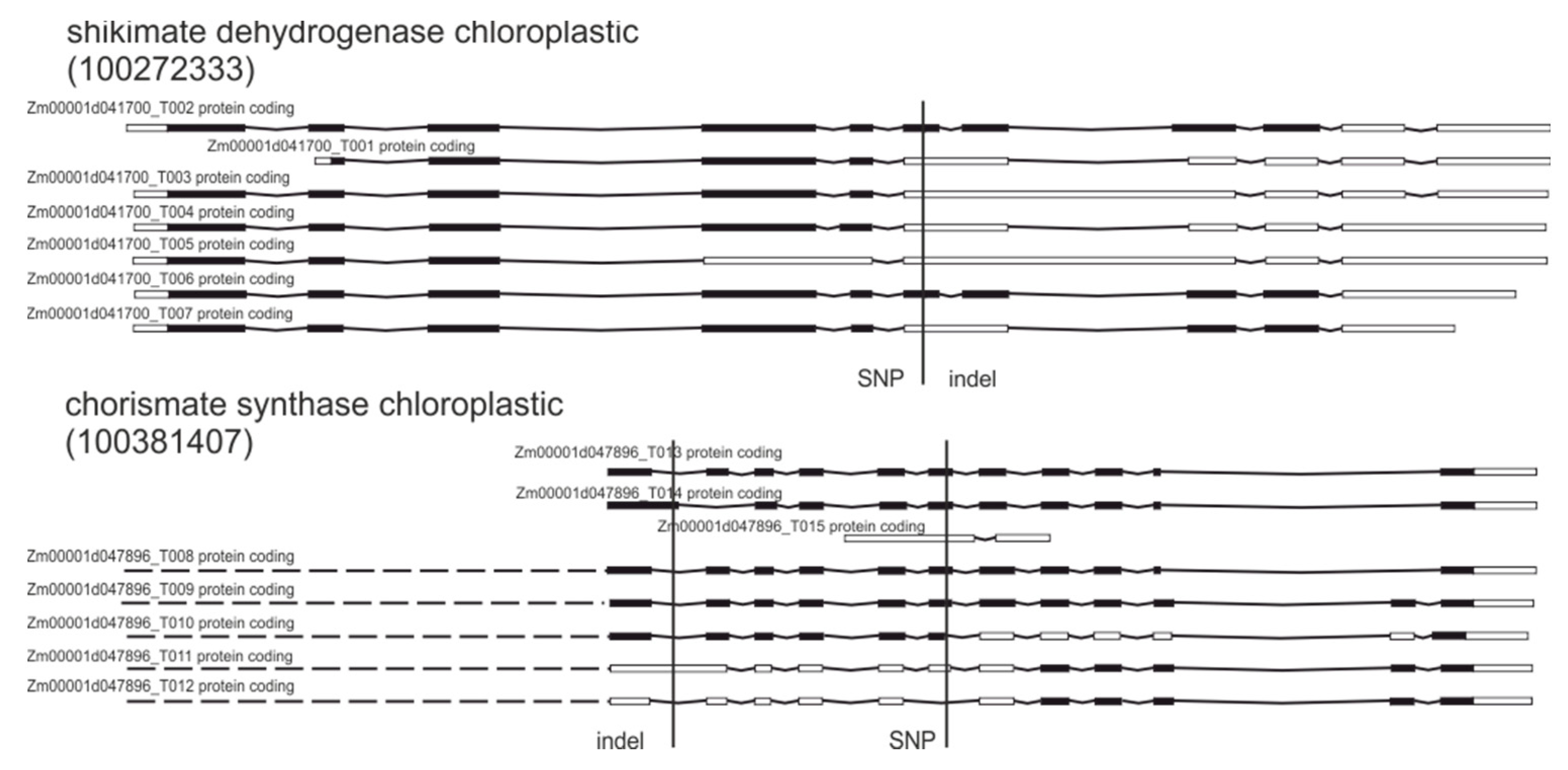
2.4. Gene Ontology Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Material and Sequencing
5.2. Preparation and Assessment of Sequencing Data
5.3. Calling Variance
5.4. Structure Variation Detection
5.5. Functional Analysis
5.6. Sequence Deposition
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Jaenicke-Despres, V.; Buckler, E.; Smith, B.D.; Gilbert, M.T.; Cooper, A.; Doebley, J.; Paabo, S. Early allelic selection in maize as revealed by ancient DNA. Science 2003, 302, 1206–1208. [Google Scholar] [CrossRef]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D.; et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2018, 115, E334–E341. [Google Scholar] [CrossRef] [PubMed]
- Tyczewska, A.; Woźniak, E.; Gracz, J.; Kuczyński, J.; Twardowski, T. Towards food security: Current state and future prospects of agrobiotechnology. Trends Biotechnol. 2018, 36, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Craine, J.M.; Dybzinski, R. Mechanisms of plant competition for nutrients, water and light. Funct. Ecol. 2013, 27, 833–840. [Google Scholar] [CrossRef]
- Żywicki, M.; Gracz, J.; Karłowski, W.; Twardowski, T.; Tyczewska, A. Expression of miRNAs involved in phosphate homeostasis and senescence is altered in glyphosate-Treated maize. Acta Physiol. Plant. 2015, 37, 265. [Google Scholar] [CrossRef]
- Heap, I.; Duke, S.O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 2018, 74, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Boocock, M.R.; Coggins, J.R. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 1983, 154, 127–133. [Google Scholar] [CrossRef]
- Denis, M.-H.; Delrot, S. Carrier-Mediated uptake of glyphosate in broad bean (Vicia faba) via a phosphate transporter. Physiol. Plant. 1993, 87, 569–575. [Google Scholar] [CrossRef]
- Geiger, D.R.; Kapitan, S.W.; Tucci, M.A. Glyphosate inhibits photosynthesis and allocation of carbon to starch in sugar beet leaves. Plant Physiol. 1986, 82, 468–472. [Google Scholar] [CrossRef]
- Hetherington, P.R.; Marshall, G.; Kirkwood, R.C.; Warner, J.M. Absorption and efflux of glyphosate by cell suspensions. J. Exp. Bot. 1998, 49, 527–533. [Google Scholar] [CrossRef]
- Morin, F.; Vera, V.; Nurit, F.; Tissut, M.; Marigo, G. Glyphosate uptake in Catharanthus roseus cells: Role of a phosphate transporter. Pestic. Biochem. Physiol. 1997, 58, 13–22. [Google Scholar] [CrossRef]
- Orcaray, L.; Zulet, A.; Zabalza, A.; Royuela, M. Impairment of carbon metabolism induced by the herbicide glyphosate. J. Plant Physiol. 2012, 169, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Servaites, J.C.; Tucci, M.A.; Geiger, D.R. Glyphosate effects on carbon assimilation, ribulose bisphosphate carboxylase activity, and metabolite levels in sugar beet leaves. Plant Physiol. 1987, 85, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Shaner, D.L. Role of translocation as a mechanism of resistance to glyphosate. Weed Sci. 2009, 57, 118–123. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.G.; Lee, K.W.; Alam, I.; Lee, S.H.; Bahk, J.D.; Lee, B.H. Glyphosate-Induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol. Biochem. 2008, 46, 1062–1070. [Google Scholar] [CrossRef]
- Das, M.; Reichman, J.R.; Haberer, G.; Welzl, G.; Aceituno, F.F.; Mader, M.T.; Watrud, L.S.; Pfleeger, T.G.; Gutierrez, R.A.; Schaffner, A.R.; et al. A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus. Plant Mol. Biol. 2010, 72, 545–556. [Google Scholar] [CrossRef]
- Unver, T.; Bakar, M.; Shearman, R.C.; Budak, H. Genome-Wide profiling and analysis of Festuca arundinacea miRNAs and transcriptomes in response to foliar glyphosate application. Mol. Genet. Genom. 2010, 283, 397–413. [Google Scholar] [CrossRef]
- Yu, W.; Zhang, R.; Li, R.; Guo, S. Isolation and characterization of glyphosate-Regulated genes in soybean seedlings. Plant Sci. 2007, 172, 497–504. [Google Scholar] [CrossRef]
- Trenkamp, S.; Eckes, P.; Busch, M.; Fernie, A.R. Temporally resolved GC-MS-based metabolic profiling of herbicide treated plants treated reveals that changes in polar primary metabolites alone can distinguish herbicides of differing mode of action. Metabolomics 2009, 5, 277–291. [Google Scholar] [CrossRef]
- Meyers, B.C.; Tingey, S.V.; Morgante, M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genom. Res. 2001, 11, 1660–1676. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-Wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.; Beridze, T.; Cantu, D.; Gaut, B.S. The population genetics of structural variants in grapevine domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, X.; Liang, P.; Tang, H. Cataloguing plant genome structural variations. Curr. Issues Mol. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-Genomics for crop improvement. Mol. Plant. 2019, 12, 156–169. [Google Scholar] [CrossRef]
- Andorf, C.M.; Cannon, E.K.; Portwood, J.L.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Braun, B.L.; Campbell, D.A.; Vinnakota, A.G.; Sribalusu, V.V.; et al. MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 2015, 44, D1195–D1201. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Forlani, G.; Racchi, M.L. Glyphosate tolerance in maize (Zea mays L.). 1. Differential response among inbred lines. Euphytica 1995, 82, 157–164. [Google Scholar]
- Alexandrov, N.N.; Brover, V.V.; Freidin, S.; Troukhan, M.E.; Tatarinova, T.V.; Zhang, H.; Swaller, T.; Lu, Y.; Bouck, J.; Flavell, R.B.; et al. Insights into corn genes derived from large-Scale cDNA sequencing. Plant Mol. Biol. 2009, 69, 179–194. [Google Scholar] [CrossRef]
- Soderlund, C.; Descour, A.; Kudrna, D.; Bomhoff, M.; Boyd, L.; Currie, J.; Angelova, A.; Collura, K.; Wissotski, M.; Ashley, E.; et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 2009, 5, e1000740. [Google Scholar] [CrossRef] [PubMed]
- Vega-Arreguin, J.C.; Ibarra-Laclette, E.; Jimenez-Moraila, B.; Martinez, O.; Vielle-Calzada, J.P.; Herrera-Estrella, L.; Herrera-Estrella, A. Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genom. 2009, 10, 299. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagen. 2008, 49, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, H.; Zhang, C.; Wei, S.; Huang, Z.; Chen, J.; Wang, X. Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 2015, 242, 859–868. [Google Scholar] [CrossRef]
- Funke, T.; Han, H.; Healy-Fried, M.L.; Fischer, M.; Schönbrunn, E. Molecular basis for the herbicide resistance of roundup ready crops. Proc. Natl. Acad. Sci. USA 2006, 103, 13010–13015. [Google Scholar] [CrossRef] [PubMed]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.; Kabsch, W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Rubin, J.L.; Gaines, G.C.; Jensen, R.A. Glyphosate inhibition of 5-enolpyruvylshikimate 3-phosphate synthase from suspension-Cultured cells of Nicotiana silvestris. Plant Physiol. 1984, 75, 839–845. [Google Scholar] [CrossRef]
- Dong, Y.; Ng, E.; Lu, J.; Fenwick, T.; Tao, Y.; Bertain, S.; Sandoval, M.; Bermudez, E.; Hou, Z.; Patten, P.; et al. Desensitizing plant EPSP synthase to glyphosate: Optimized global sequence context accommodates a glycine-To-Alanine change in the active site. J. Biol. Chem. 2019, 294, 716–725. [Google Scholar] [CrossRef]
- Zhou, M.; Xu, H.; Wei, X.; Ye, Z.; Wei, L.; Gong, W.; Wang, Y.; Zhu, Z. Identification of a glyphosate-Resistant mutant of rice 5-enolpyruvylshikimate 3-phosphate synthase using a directed evolution strategy. Plant Physiol. 2006, 140, 184–195. [Google Scholar] [CrossRef]
- Kahrizi, D.; Salmanian, A.H.; Afshari, A.; Moieni, A.; Mousavi, A. Simultaneous substitution of Gly96 to Ala and Ala183 to Thr in 5-enolpyruvylshikimate-3-phosphate synthase gene of E. coli (k12) and transformation of rapeseed (Brassica napus L.) in order to make tolerance to glyphosate. Plant Cell Rep. 2007, 26, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, Y.; Zhang, C.; Jiang, C.; Huang, H.; Wei, S. Molecular basis of natural tolerance to glyphosate in Convolvulus arvensis. Sci. Rep. 2019, 9, 8133. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, C.; Huang, H.; Wei, S.; Huang, Z.; Wang, H.; Zhao, D.; Zhang, C. Characterization of Eleusine indica with gene mutation or amplification in EPSPS to glyphosate. Pestic Biochem. Physiol. 2017, 143, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Cao, Y.P. Improvement of glyphosate resistance through concurrent mutations in three amino acids of the Pantoea sp. 5-enolpyruvylshikimate-3-phosphate synthase. J. Microbiol. Biotechnol. 2018, 28, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G.; Aharoni, A. Shikimate Pathway and Aromatic Amino Acid Biosynthesis; eLS. John Wiley & Sons, Ltd.: Chichester, UK, 2012. [Google Scholar] [CrossRef]
- Gomes, M.P.; Le Manac’h, S.G.; Moingt, M.; Smedbol, E.; Paquet, S.; Labrecque, M.; Lucotte, M.; Juneau, P. Impact of phosphate on glyphosate uptake and toxicity in willow. J. Hazard. Mater. 2016, 304, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lv, S.; Jiang, P.; Li, Y. Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 2017, 8, 817. [Google Scholar] [CrossRef]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Pochon, N.; Ayadi, A.; Nakanishi, T.M.; Thibaud, M. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef]
- Shi, J.; Yi, K.; Liu, Y.; Xie, L.; Zhou, Z.; Chen, Y.; Hu, Z.; Zheng, T.; Liu, R.; Chen, Y.; et al. Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiology 2015, 167, 671–681. [Google Scholar] [CrossRef]
- O′Leary, B.; Rao, S.K.; Kim, J.; Plaxton, W.C. Bacterial-Type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class-2 PEPC complex of vascular plants. J. Biol. Chem. 2009, 284, 24797–24805. [Google Scholar] [CrossRef]
- Streatfield, S.J.; Weber, A.; Kinsman, E.A.; Häusler, R.E.; Li, J.; Post-Beittenmiller, D.; Kaiser, W.M.; Pyke, K.A.; Flügge, U.I.; Chory, J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-Dependent nuclear gene expression. Plant Cell 1999, 11, 1609–1621. [Google Scholar] [CrossRef]
- Masumoto, C.; Miyazawa, S.; Ohkawa, H.; Fukuda, T.; Taniguchi, Y.; Murayama, S.; Kusano, M.; Saito, K.; Fukayama, H.; Miyao, M. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc. Natl. Acad. Sci. USA 2010, 107, 5226–5231. [Google Scholar] [CrossRef] [PubMed]
- De María, N.; Becerril, J.M.; García-Plazaola, J.I.; Hernandez, A.; De Felipe, M.R.; Fernandez-Pascual, M. New insights on glyphosate mode of action in nodular metabolism: Role of shikimate accumulation. J. Agric Food Chem. 2006, 54, 2621–2628. [Google Scholar] [CrossRef] [PubMed]
- Lu, M. Structures of multidrug and toxic compound extrusion transporters and their mechanistic implications. Channels (Austin) 2016, 10, 88–100. [Google Scholar] [CrossRef]
- Takanashi, K.; Shitan, N.; Yazaki, K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 2014, 31, 417–430. [Google Scholar] [CrossRef]
- Remy, E.; Duque, P. Beyond cellular detoxification: A plethora of physiological roles for MDR transporter homologs in plants. Front Physiol. 2014, 5, 201. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Magwanga, R.O.; Guo, X.; Kirungu, J.N.; Lu, H.; Cai, X.; Zhou, Z.; Wei, Y.; Wang, X.; Zhang, Z.; et al. Genome-Wide Analysis of Multidrug and Toxic Compound Extrusion (MATE) Family in Gossypium raimondii and Gossypium arboreum and Its Expression Analysis Under Salt, Cadmium, and Drought Stress. G3 (Bethesda) 2018, 8, 2483–2500. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Mahmoud, M.; Zywicki, M.; Twardowski, T.; Karlowski, W.M. Efficiency of PacBio long read correction by 2nd generation Illumina sequencing. Genomics 2019, 111, 43–49. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-Generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-Molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef]
- English, A.C.; Salerno, W.J.; Reid, J.G. PBHoney: Identifying genomic variants via long-Read discordance and interrupted mapping. BMC Bioinform. 2014, 15, 180. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]


| Tolerant Line (S245) | Sensitive Line (S79757) | |||||
|---|---|---|---|---|---|---|
| Library Size | Millions of Reads | Genome Coverage | Alignment Rate | Millions of Reads | Genome Coverage | Alignment Rate |
| 400 bp | 816 | 37.5 | 97.1% | 821 | 37.7 | 96.6% |
| 500 bp | 664 | 27.8 | 85.3% | 615 | 29.1 | 97.0% |
| 8 kb | 129 | 3.7 | 86.4% | 74 | 3.0 | 87.7% |
| 11 kb | 59 | 2.2 | 86.0% | 116 | 2.6 | 70.0% |
| Variant Group | SNPs | Indels |
|---|---|---|
| All | 13,778,463 | 2,443,262 |
| Tolerant line-specific | 4,068,829 | 729,866 |
| Located in coding sequence | 113,775 | 15,277 |
| Gene | Protein | Variation | Consequence |
|---|---|---|---|
| 100272333 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase chloroplastic | SNPs, indels | Splice donor/frame shift |
| 100381407 | Chorismate synthase chloroplastic | SNPs, indels | Splice donor/frame shift |
| Gene | Protein | Variation | Consequence |
|---|---|---|---|
| Zm00001d051945 | Phosphate transporter 2 | SNP | Splice acceptor |
| Zm00001d018445 | Phosphate transporter 3 | indel | Splice donor |
| Zm00001d012747 | Putative sugar phosphate/phosphate translocator | SNP, indel | Splice donor/Splice acceptor |
| Zm00001d021653 | Glucose-6-phosphate/phosphate translocator 2 | indel | Frame shift |
| 100191756 | Probable sugar phosphate/phosphate translocator | indel | Frame shift |
| Zm00001d011388 | Putative sugar phosphate/phosphate translocator | indel | Frame shift |
| Gene | Protein | Variation | Consequence |
|---|---|---|---|
| 100283648 | Phosphoenolpyruvate/phosphate translocator 1 chloroplastic | SNP | Splice acceptor |
| 103649694 | Phosphoenolpyruvate/phosphate translocator 2 chloroplastic | indels | Frame shift |
| Zm00001d044715 | Phosphoenolpyruvate/phosphate translocator 2 chloroplastic | indel | Frame shift |
| Zm00001d037659 | Phosphoenolpyruvate/phosphate translocator 2 chloroplastic | SV insertion | Stop lost |
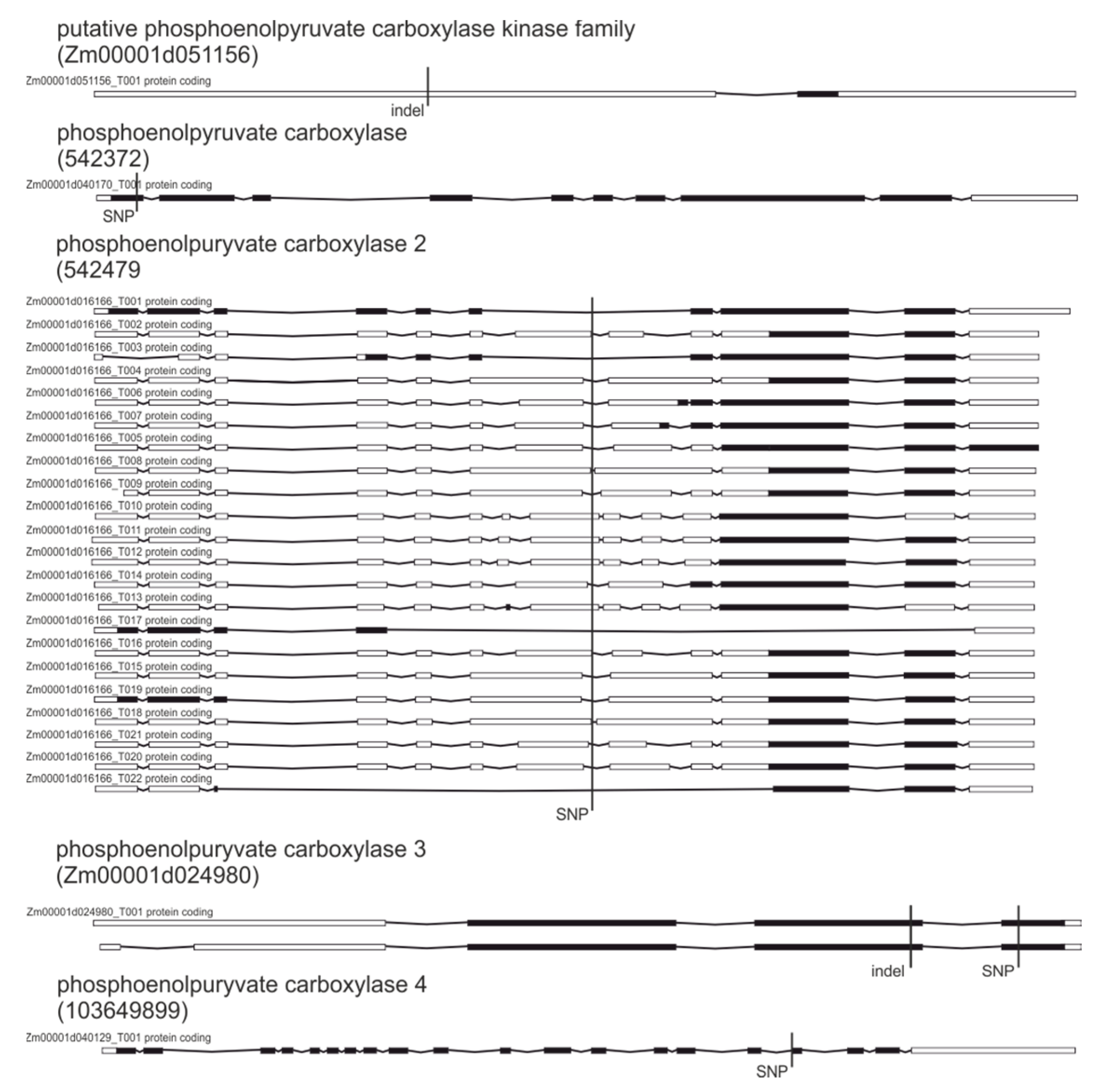
| 542372 | Phosphoenolpyruvate carboxylase | SNP | Stop gained |
| Zm00001d053453 | Phosphoenolpyruvate carboxylase isoform 1 | SNPs, indels | Splice acceptor/Splice donor |
| 542479 | Phosphoenolpyruvate carboxylase 2 | SNP | Splice donor |
| Zm00001d024980 | Phosphoenolpyruvate carboxylase 3 | SNPs, indels | Stop gained/frame shift |
| 103649899 | Phosphoenolpyruvate carboxylase 4 | SNP | Splice acceptor |
| Zm00001d051156 | Putative phosphoenolpyruvate carboxylase kinase family protein | SNPs, indel | Stop gained/frame shift |
| Gene | Protein | Variation | Consequence |
|---|---|---|---|
| Zm00001d005080 | Multidrug and toxic compound extrusion5 | SNPs, indels | Splice acceptor/frame shift |
| Zm00001d013810 | Multidrug and toxic compound extrusion2 | indels, structural variant (SV) insertion | Frame shift/ start loss |
| 100383875 | Multidrug and toxic compound extrusion3 | indel | Frame shift |
| 100193278 | Multidrug and toxic compound extrusion4 | indel | Frame shift |
| Zm00001d035115 | Multidrug and toxic compound extrusion1 | indel | Stop gained |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoud, M.; Gracz-Bernaciak, J.; Żywicki, M.; Karłowski, W.; Twardowski, T.; Tyczewska, A. Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance. Plants 2020, 9, 523. https://doi.org/10.3390/plants9040523
Mahmoud M, Gracz-Bernaciak J, Żywicki M, Karłowski W, Twardowski T, Tyczewska A. Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance. Plants. 2020; 9(4):523. https://doi.org/10.3390/plants9040523
Chicago/Turabian StyleMahmoud, Medhat, Joanna Gracz-Bernaciak, Marek Żywicki, Wojciech Karłowski, Tomasz Twardowski, and Agata Tyczewska. 2020. "Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance" Plants 9, no. 4: 523. https://doi.org/10.3390/plants9040523
APA StyleMahmoud, M., Gracz-Bernaciak, J., Żywicki, M., Karłowski, W., Twardowski, T., & Tyczewska, A. (2020). Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance. Plants, 9(4), 523. https://doi.org/10.3390/plants9040523





