Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis
Abstract
:1. Background
2. Results
2.1. De Novo Assembly of the Jojoba Transcriptome
2.2. Abundance Estimation
2.3. ORF Prediction
2.4. Annotation
2.5. Annotation on GO Database
2.6. EggNOG Analysis
2.7. Gene Expression Analysis
2.7.1. Gene Expression of Lipid Biosynthesis Genes
2.7.2. Fatty Acid Biosynthesis
2.7.3. Fatty Acid Desaturation
2.7.4. Fatty acid Elongation
2.7.5. Fatty Alcohol Synthesis and Oxidation
2.7.6. Triacylglycerol (TGA) Metabolism
2.7.7. Phospholipid Metabolism
2.7.8. Wax ester Biosynthesis
2.7.9. Lipid Transfer and Storage Proteins/Enzymes
3. Discussion
3.1. Lipid Biosynthesis Gene Expression Profiling
3.2. Fatty Acid Biosynthesis
3.3. Fatty Acid Desaturation
3.4. Fatty Acid Elongation
3.5. LCFA Biosynthesis
3.6. Triglyceride (TGA) Metabolism
3.7. Phospholipid Metabolism
3.8. Wax Ester Biosynthesis
3.9. Lipid Transfer and Storage Proteins/Enzymes
4. Materials and Methods
4.1. Purification of mRNA
4.2. Synthesis of cDNA Libraries
4.3. Sequencing and Sequence Analysis
4.4. ORF Prediction and Abundance Estimation
4.5. Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Harry-O’kuru, R.E.; Biresaw, G.; Gordon, S.; Xu, J. Physical Characteristics of Tetrahydroxy and Acylated Derivatives of Jojoba Liquid Wax in Lubricant Applications. J. Anal. Methods Chem. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miklaszewska, M.; Banaś, A. Biochemical characterization and substrate specificity of jojoba fatty acyl-CoA reductase and jojoba wax synthase. Plant. Sci. 2016, 249, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Kharb, P.; Mitra, C.; Uppal, S. Early diagnosis of sex in jojoba, Simmondsia chinensis (link) schneider by sequence characterized amplified region marker. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2014, 84, 251–255. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Bayoumi, S.A.L.H.; Salama, A.A.R.; Salem-Bekhit, M.M.; Abd-Alrahman, S.H.; Sayed, H.M. Antioxidant lipoxygenase inhibitors from the leaf extracts of Simmondsia chinensis. Asian Pac. J. Trop. Med. 2014, 7, S521–S526. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Röttig, A.; Steinbüchel, A. Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 277–321. [Google Scholar] [CrossRef] [Green Version]
- Sandha, G.; Swami, V. Jojoba oil as an organic, shelf stable standard oil-phase base for cosmetic industry. Rasayan J. Chem. 2009, 2, 300–306. [Google Scholar]
- Bouaid, A.; Bajo, L.; Martinez, M.; Aracil, J. Optimization of biodiesel production from jojoba oil. Process. Saf. Environ. Prot. 2007, 85, 378–382. [Google Scholar] [CrossRef]
- Araiza-Lizarde, N.; Alcaraz-Meléndez, L.; Angulo-Escalante, M.A.; Reynoso-Granados, T.; Cruz-Hernández, P.; Calderón-Vázquez, C.L. Physicochemical composition of seed oil of wild jojoba populations in northwestern Mexico. J. Food Nutr. Res. 2017, 5, 443–450. [Google Scholar]
- Al-Obaidi, J.R.; Halabi, M.F.; AlKhalifah, N.S.; Asanar, S.; Al-Soqeer, A.A.; Attia, M. A review on plant importance, biotechnological aspects, and cultivation challenges of jojoba plant. Biol. Res. 2017, 50, 25. [Google Scholar] [CrossRef] [Green Version]
- Habashy, R.R.; Abdel-Naim, A.B.; Khalifa, A.E.; Al-Azizi, M.M. Anti-inflammatory effects of jojoba liquid wax in experimental models. Pharmacol. Res. 2005, 51, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mageed, W.M.; Bayoumi, S.A.; Al-Wahaibi, L.H.; Li, L.; Sayed, H.M.; Abdelkader, M.S.; El-Gamal, A.A.; Liu, M.; Zhang, J.; Zhang, L.; et al. Noncyanogenic cyanoglucoside cyclooxygenase inhibitors from Simmondsia chinensis. Org. Lett. 2016, 18, 1728–1731. [Google Scholar] [CrossRef] [PubMed]
- Bouali, A.; Bellirou, A.; Boukhatem, N.; Hamal, A.; Bouammali, B. Enzymatic detoxification of jojoba meal and effect of the resulting meal on food intake in rats. Nat. Prod. Res. 2008, 22, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Hills, G. Industrial use of lipases to produce fatty acid esters. Eur. J. Lipid Sci. Technol. 2003, 105, 601–607. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Stöveken, T.; Luftmann, H.; Malkus, U.; Reichelt, R.; Steinbüchel, A. Neutral lipid biosynthesis in engineered Escherichia coli: Jojoba oil-like wax esters and fatty acid butyl esters. Appl Environ. Microbiol. 2006, 72, 1373–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, S.N.; Sharma, B.K.; Moser, B.R. Preparation of biofuel using acetylatation of jojoba fatty alcohols and assessment as a blend component in ultralow sulfur diesel fuel. Energy Fuels 2010, 24, 3189–3194. [Google Scholar] [CrossRef]
- Schei, M.L. Production of Biodiesel from Jojoba Oil with Calcium Glyceoxide as Catalyst; Norwegian University of Life Sciences: Åslund, Norway, 2017. [Google Scholar]
- Sandouqa, A.; Al-Hamamre, Z. Energy analysis of biodiesel production from jojoba seed oil. Renew. Energy 2019, 130, 831–842. [Google Scholar] [CrossRef]
- Bao, Y.Z.; Yao, Z.Q.; Cao, X.L.; Peng, J.F.; Xu, Y.; Chen, M.X.; Zhao, S.F. Transcriptome analysis of Phelipanche aegyptiaca seed germination mechanisms stimulated by fluridone, TIS108, and GR24. PLoS ONE 2017, 12, e0187539. [Google Scholar] [CrossRef] [Green Version]
- Sreeharsha, R.V.; Mudalkar, S.; Singha, K.T.; Reddy, A.R. Unravelling molecular mechanisms from floral initiation to lipid biosynthesis in a promising biofuel tree species, Pongamia pinnata using transcriptome analysis. Sci. Rep. 2016, 6, 34315. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ni, J.; Shah, F.A.; Wang, Q.; Wang, Z.; Wu, L.; Fu, S. Transcriptome analysis of pecan seeds at different developing stages and identification of key genes involved in lipid metabolism. PLoS ONE 2018, 13, e0195913. [Google Scholar] [CrossRef]
- Abdullah, H.M.; Akbari, P.; Paulose, B.; Schnell, D.; Qi, W.; Park, Y.; Pareek, A.; Dhankher, O.P. Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol. Biofuels 2016, 9, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; He, Z.; Yin, Y.; Xu, X.; Wu, W.; Li, L. Transcriptome sequencing and analysis during seed growth and development in Euryale ferox Salisb. BMC Genom. 2018, 19, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Hao, X.; Jin, Y.; Guo, X.; Shao, Q.; Kumar, K.S.; Ahlawat, Y.K.; Harry, D.E.; Joshi, C.P.; Zheng, Y. Temporal transcriptome profiling of developing seeds reveals a concerted gene regulation in relation to oil accumulation in Pongamia (Millettia pinnata). BMC Plant. Biol. 2018, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wang, K.; Zhou, C.; Xie, Y.; Yao, X.; Yin, H. Seed transcriptomics analysis in Camellia oleifera uncovers genes associated with oil content and fatty acid composition. Int. J. Mol. Sci. 2018, 19, 118. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Yu, R.; Sun, D.; Rahman, M.; Xie, L.; Hu, J.; He, L.; Kilaru, A.; Niu, L.; Zhang, Y. Comparative Transcriptome Analysis Reveals an Efficient Mechanism of α-Linolenic Acid in Tree Peony Seeds. Int. J. Mol. Sci. 2019, 20, 65. [Google Scholar] [CrossRef] [Green Version]
- Liao, B.; Hao, Y.; Lu, J.; Bai, H.; Guan, L.; Zhang, T. Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids. BMC Genom. 2018, 19, 213. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; Agrawal, V.; Gupta, S.; Kumar, R.; Prasad, M. ISSR marker-assisted selection of male and female plants in a promising dioecious crop: Jojoba (Simmondsia chinensis). Plant. Biotechnol. Rep. 2008, 2, 239–243. [Google Scholar] [CrossRef]
- Heikrujam, M.; Sharma, K.; Kumar, J.; Agrawal, V. Generation and validation of unique male sex-specific sequence tagged sites (STS) marker from diverse genotypes of dioecious Jojoba-Simmondsia chinensis (Link) Schneider. Euphytica 2014, 199, 363–372. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Development of sex-linked AFLP markers in Simmondsia chinensis. Plant. Breed. 2011, 130, 114–116. [Google Scholar] [CrossRef]
- Heikrujam, M.; Kumar, J.; Agrawal, V. Genetic diversity analysis among male and female Jojoba genotypes employing gene targeted molecular markers, start codon targeted (SCoT) polymorphism and CAAT box-derived polymorphism (CBDP) markers. Meta Gene 2015, 5, 90–97. [Google Scholar] [CrossRef]
- Al-Obaidi, J.R.; Rahmad, N.; Hanafi, N.M.; Halabi, M.F.; Al-Soqeer, A.A. Comparative proteomic analysis of male and female plants in jojoba (Simmondsia chinensis) leaves revealed changes in proteins involved in photosynthesis, metabolism, energy, and biotic and abiotic stresses. Acta Physiol. Plant. 2017, 39, 179. [Google Scholar] [CrossRef]
- Metz, J.G.; Pollard, M.R.; Anderson, L.; Hayes, T.R.; Lassner, M.W. Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 2000, 122, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, D.J. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 2001, 40, 325–438. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant. Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, N.; Moriyama, T.; Toyoshima, M.; Sato, N. Construction of global acyl lipid metabolic map by comparative genomics and subcellular localization analysis in the red alga Cyanidioschyzon merolae. Front. Plant. Sci. 2016, 7, 958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haslam, T.M.; Kunst, L. Extending the story of very-long-chain fatty acid elongation. Plant. Sci. 2013, 210, 93–107. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.; Arondel, V.; Bates, P. Acyl-lipid metabolism. Arab B 2013, 11, e0161. [Google Scholar] [CrossRef] [Green Version]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Baud, S.; Lepiniec, L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 2009, 47, 448–455. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Kilaru, A.; Cao, X.; Durrett, T.P.; Fan, J.; Jensen, J.K.; Thrower, N.A.; Pauly, M.; Wilkerson, C.; Ohlrogge, J.B. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011, 68, 1014–1027. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Chye, M.-L. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 2011, 50, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tan, R.-K.; Guo, X.-J.; Fu, Z.-L.; Wang, Z.; Zhang, Z.-Y.; Tan, X.L. Transcriptome analysis comparison of lipid biosynthesis in the leaves and developing seeds of Brassica napus. PLoS ONE 2015, 10, e0126250. [Google Scholar] [CrossRef]
- Kim, J.; Jung, J.H.; Lee, S.B.; Go, Y.S.; Kim, H.J.; Cahoon, R.; Markham, J.E.; Cahoon, E.B.; Suh, M.C. Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 2013, 162, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, M.; Sun, Y.; Wang, Y.; Li, T.; Chai, G.; Jiang, W.; Shan, L.; Li, C.; Xiao, E.; et al. FAR5, a fatty acyl-coenzyme A reductase, is involved in primary alcohol biosynthesis of the leaf blade cuticular wax in wheat (Triticum aestivum L.). J. Exp. Bot. 2014, 66, 1165–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justin, A.M.; Kader, J.C.; Collin, S. Phosphatidylinositol synthesis and exchange of the inositol head are catalysed by the single phosphatidylinositol synthase 1 from Arabidopsis. Eur. J. Biochem. 2002, 269, 2347–2352. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Yu, Y.; Mizoi, J.; Fujiki, Y.; Saito, K.; Nishijima, M.; Lee, Y.; Nishida, I. PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J. 2011, 67, 648–661. [Google Scholar] [CrossRef]
- Faure, L.; Coulon, D.; Laroche-Traineau, J.; Le Guedard, M.; Schmitter, J.-M.; Testet, E.; Lessire, R.; Bessoule, J. Discovery and characterization of an Arabidopsis thaliana N-acylphosphatidylethanolamine synthase. J. Biol. Chem. 2009, 284, 18734–18741. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Peisker, H.; Weth, A.; Baumgartner, W.; Dörmann, P.; Frentzen, M. Extraplastidial cytidinediphosphate diacylglycerol synthase activity is required for vegetative development in Arabidopsis thaliana. Plant J. 2013, 75, 867–879. [Google Scholar] [CrossRef]
- Kato, T.; Morita, M.T.; Fukaki, H.; Yamauchi, Y.; Uehara, M.; Niihama, M.; Tasaka, M. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant. Cell 2002, 14, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhou, Y.; Chen, J.; Shi, J.; Zhao, H.; Zhao, H.; Song, W.; Zhang, M.; Cui, Y.; Dong, X.; et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 2018, 50, 1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardizabal, K.D.; Metz, J.G.; Sakamoto, T.; Hutton, W.C.; Pollard, M.R.; Lassner, M.W. Purification of a jojoba embryo wax synthase, cloning of its cDNA, and production of high levels of wax in seeds of transgenic Arabidopsis. Plant Physiol. 2000, 122, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 2014, 152, 32–42. [Google Scholar] [CrossRef]
- Kader, J.-C. Lipid-transfer proteins: A puzzling family of plant proteins. Trends Plant Sci. 1997, 2, 66–70. [Google Scholar] [CrossRef]
- Zottich, U.; Da Cunha, M.; Carvalho, A.O.; Dias, G.B.; Silva, N.C.; Santos, I.S.; do Nacimento, V.V.; Miguel, E.C.; Machado, O.L.; Gomes, V.M. Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with α-amylase inhibitor properties. Biochim. Biophys. Acta (BBA) Gen. Subj. 2011, 1810, 375–383. [Google Scholar] [CrossRef]
- D’Agostino, N.; Buonanno, M.; Ayoub, J.; Barone, A.; Monti, S.M.; Rigano, M.M. Identification of non-specific lipid transfer protein gene family members in Solanum lycopersicum and insights into the features of sola l 3 protein. Sci. Rep. 2019, 9, 1607. [Google Scholar] [CrossRef]
- Asero, R.; Mistrello, G.; Roncarolo, D.; de Vries, S.C.; Gautier, M.-F.; Ciurana, C.L.; Verbeek, E.; ·Mohammadi, T.; Knul-Brettlova, V.; ·Akkerdaas, J.H.; et al. Lipid transfer protein: A pan-allergen in plant-derived foods that is highly resistant to pepsin digestion. Int. Arch. Allergy Immunol. 2001, 124, 67–69. [Google Scholar] [CrossRef]
- Finkina, E.; Melnikova, D.; Bogdanov, I. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar] [CrossRef]
- Kader, J.-C. Lipid-transfer proteins in plants. Annu. Rev. Plant Biol. 1996, 47, 627–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkina, E.; NMelnikova, D.; VBogdanov, I.; VOvchinnikova, T. Plant pathogenesis-related proteins PR-10 and PR-14 as components of innate immunity system and ubiquitous allergens. Curr. Med. Chem. 2017, 24, 1772–1787. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.; Williams, H.E.; Cannane, A. Improved gapped alignment in BLAST. IEEE/ACM Trans. Comput. Biol. Bioinform. 2004, 1, 116–129. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59. [Google Scholar] [CrossRef]
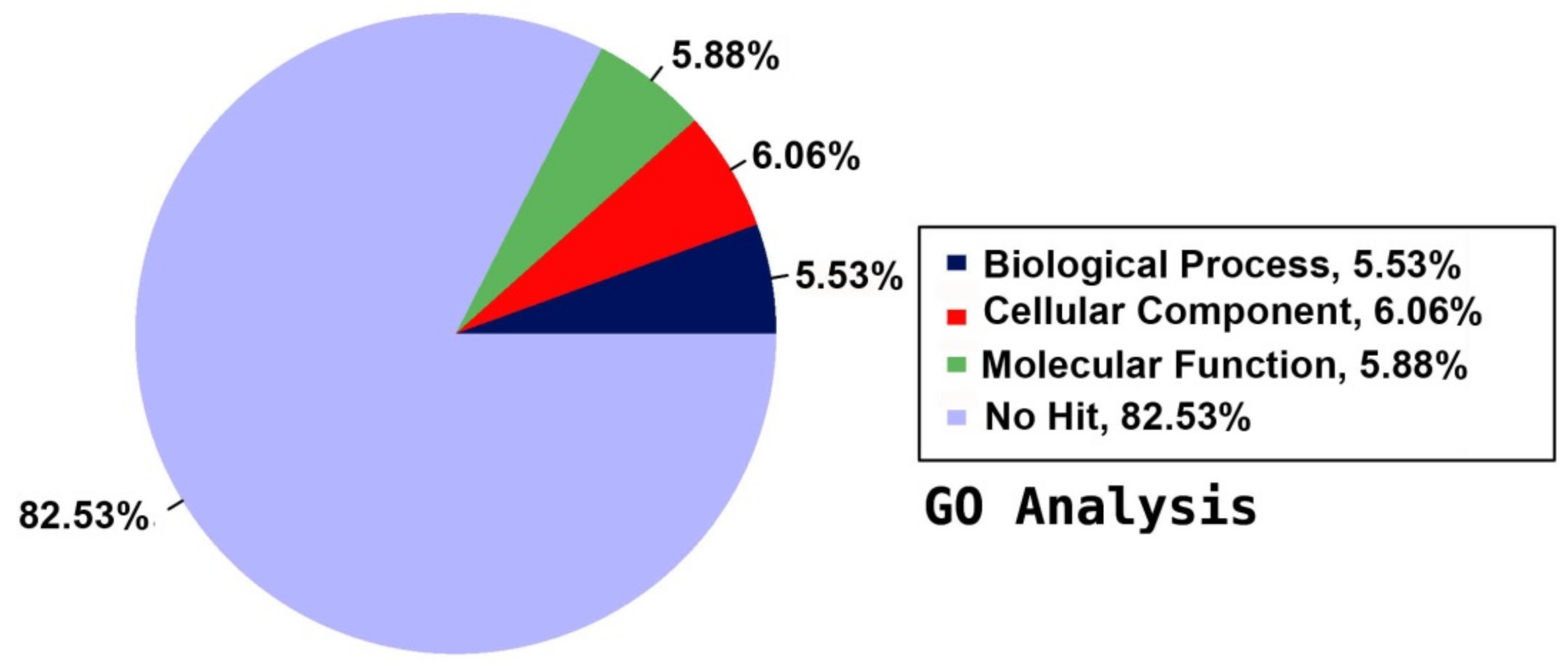
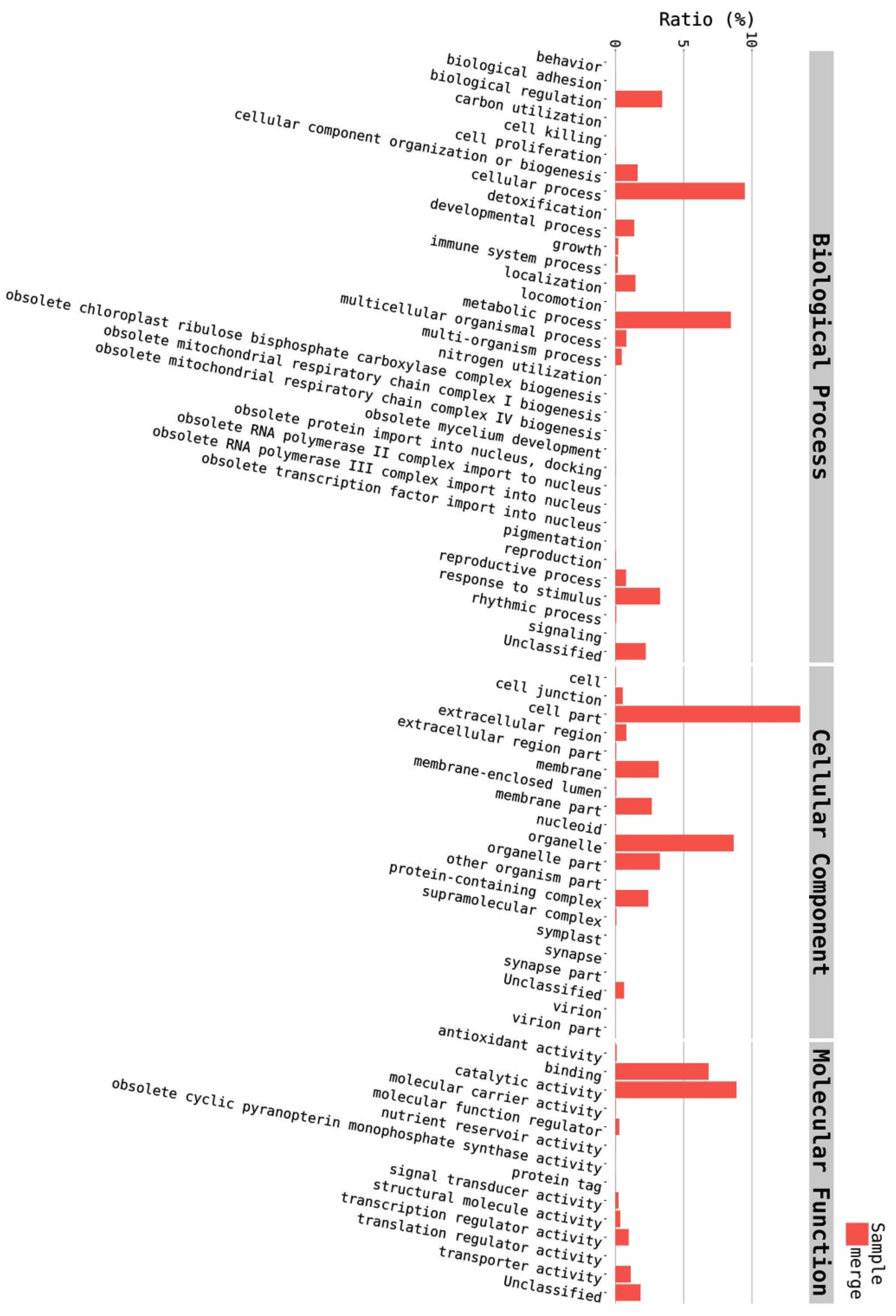
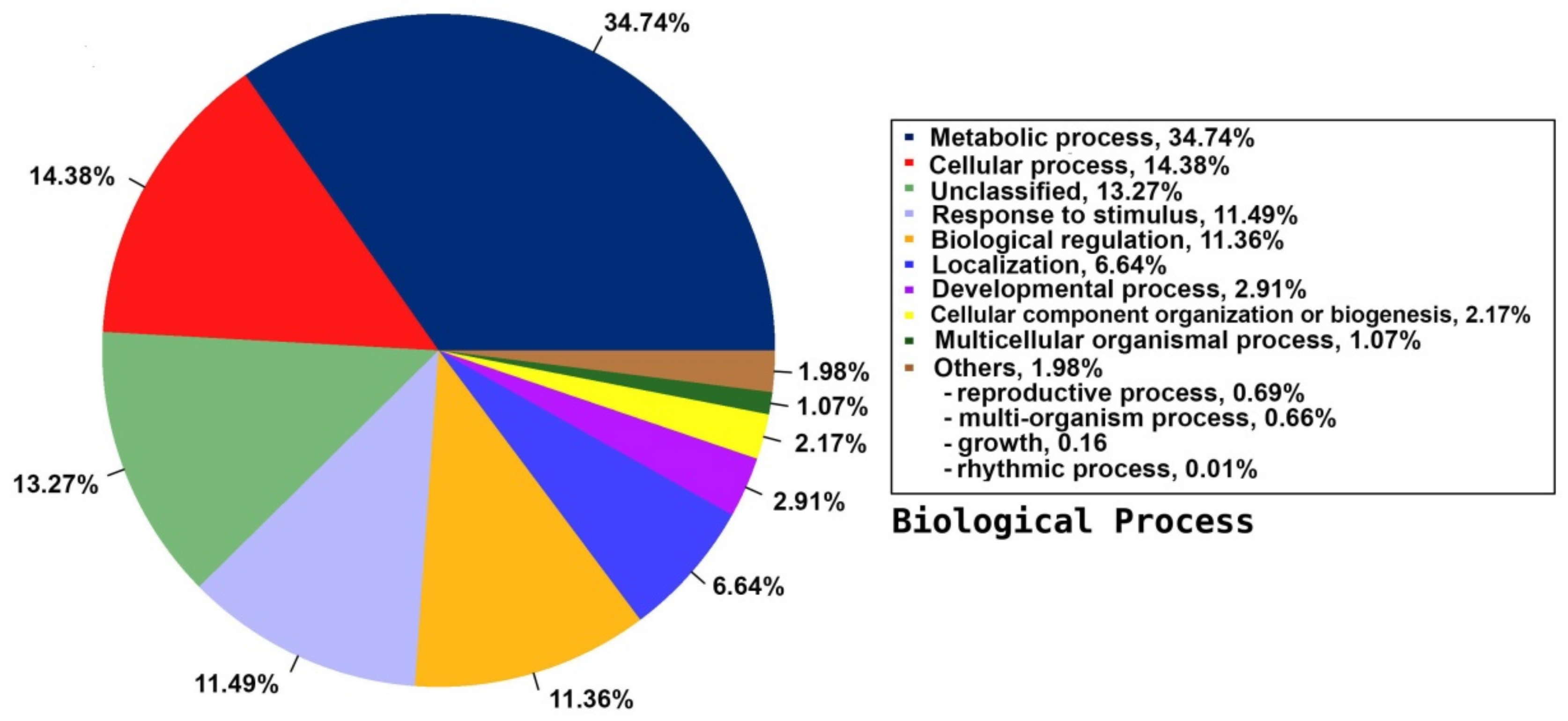

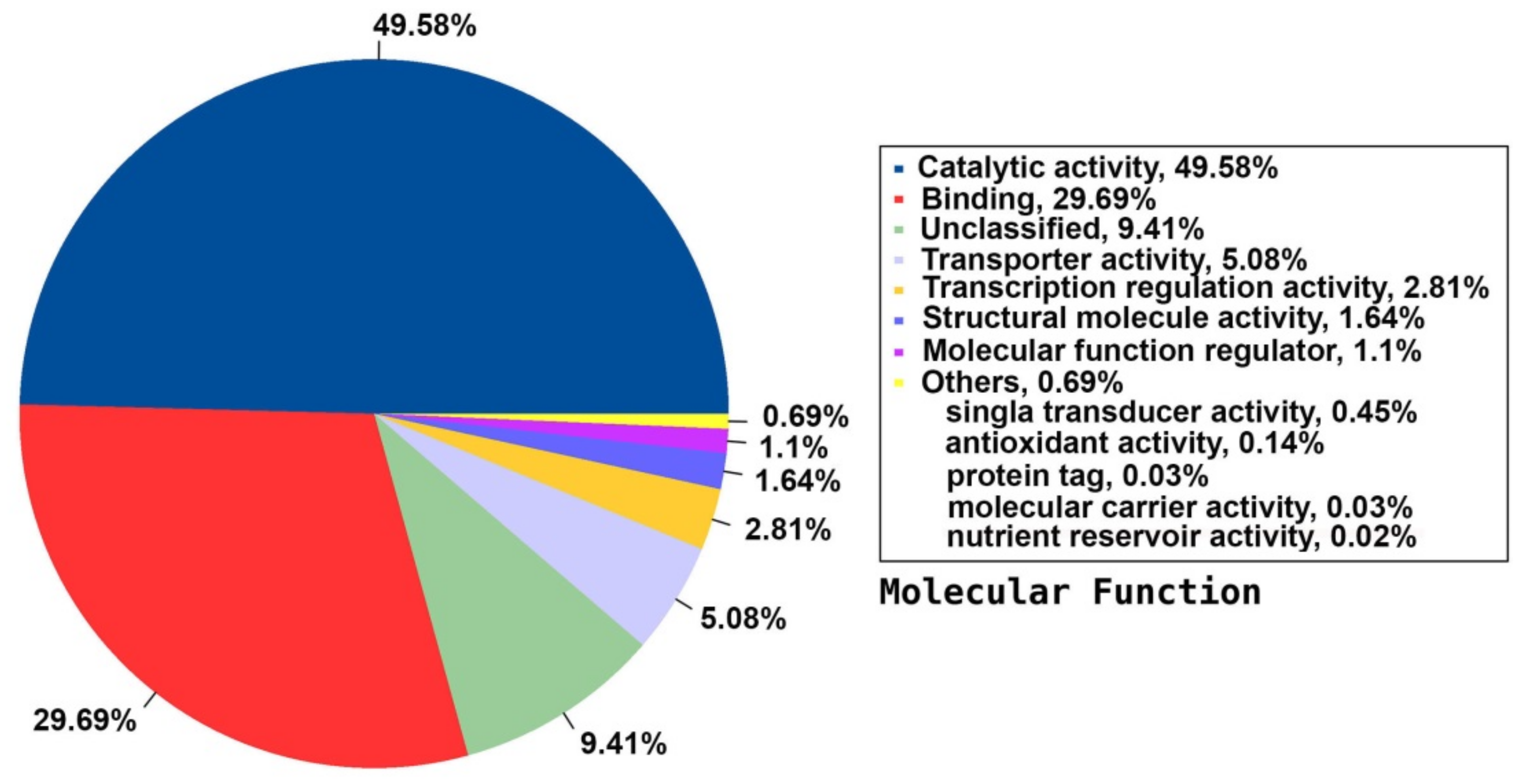


| Category | Count | Ratio | Name |
|---|---|---|---|
| Information storage and processing | |||
| J | 797 | 2.19% | Translation, ribosomal structure and biogenesis |
| A | 156 | 0.43% | RNA processing and modification |
| K | 1462 | 4.03% | Transcription |
| L | 7802 | 21.48% | Replication, recombination and repair |
| B | 152 | 0.42% | Chromatin structure and dynamics |
| Cellular processes and signaling | |||
| D | 229 | 0.63% | Cell cycle control, cell division, chromosome partitioning |
| Y | 1 | 0.0% | Nuclear structure |
| V | 284 | 0.78% | Defense mechanisms |
| T | 1542 | 4.25% | Signal transduction mechanisms |
| M | 317 | 0.87% | Cell wall/membrane/envelope biogenesis |
| N | 6 | 0.02% | Cell motility |
| Z | 274 | 0.75% | Cytoskeleton |
| W | 0 | 0% | Extracellular structures |
| U | 866 | 2.38% | Intracellular trafficking, secretion, and vesicular transport |
| O | 1612 | 4.44% | Posttranslational modification, protein turnover, chaperones |
| Metabolism | |||
| C | 504 | 1.39% | Energy production and conversion |
| G | 1031 | 2.84% | Carbohydrate transport and metabolism |
| E | 778 | 2.14% | Amino acid transport and metabolism |
| F | 183 | 0.5% | Nucleotide transport and metabolism |
| H | 211 | 0.58% | Coenzyme transport and metabolism |
| I | 390 | 1.07% | Lipid transport and metabolism |
| P | 407 | 1.12% | Inorganic ion transport and metabolism |
| Q | 446 | 1.23% | Secondary metabolites biosynthesis, transport and catabolism |
| Poorly characterized | |||
| R | 0 | 0% | General function prediction only |
| S | 16,866 | 46.44% | Function unknown |
| Contig | Read Count | FPKM | Enzyme Name | Abbreviation | Location |
|---|---|---|---|---|---|
| Fatty acid biosynthesis | |||||
| c85820_g3_i2 | 7804 | 138.28 | malonate-CoA ligase | MCAL | Cyt |
| c106491_g2_i1 | 18,389 | 88.75 | acetyl-CoA carboxylase | ACCase- α | Pt |
| c106398_g2_i1 | 9264 | 195.99 | biotin carboxylase 2 | BC | Pt |
| c105364_g14_i1 | 6138 | 229.71 | biotin carboxyl carrier protein of acetyl-CoA carboxylase | BCCP | Pt |
| c100625_g1_i2 | 178 | 4.95 | acetyl-CoA carboxylase beta subunit | ACCase- β | Pt |
| c103501_g4_i1 | 6386 | 152.71 | malonyl CoA-acyl carrier protein transacylase | MCAAT | Pt |
| c90813_g1_i1 | 13,507 | 264.77 | 3-oxoacyl-[acyl-carrier-protein] synthase I | KAS I | Pt |
| c88978_g1_i1 | 7348 | 100.10 | 3-oxoacyl-[acyl-carrier-protein] synthase II | KAS II | Pt |
| c93468_g1_i1 | 10,261 | 198.16 | 3-oxoacyl-[acyl-carrier-protein] reductase | KAR4 | Pt |
| c89737_g1_i1 | 7589 | 254.27 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | HAD | Pt |
| c92699_g1_i1 | 10,393 | 174.01 | enoyl-[acyl-carrier-protein] reductase | EAR | Pt |
| c102110_g4_i1 | 492 | 21.47 | palmitoyl-acyl carrier protein thioesterase | AAT(FATB) | Pt |
| c97538_g2_i1 | 7486 | 95.33 | long chain acyl-CoA synthetase | LACS | Pt |
| Fatty acid desaturation | |||||
| c101312_g2_i1 | 3494 | 69.07 | delta(12)-acyl-lipid-desaturase | FAD2 | ER |
| c95630_g1_i2 | 755 | 19.88 | omega-6 fatty acid desaturase | FAD6 | ER |
| c101742_g2_i1 | 1478 | 33.31 | delta(8)-fatty-acid desaturase 2 | SLD | ER |
| c100587_g1_i2 | 566 | 8.87 | sphingolipid delta(4)-desaturase DES1 | SLD | ER |
| c93084_g1_i1 | 1527 | 50.69 | delta(7)-sterol-C5(6)-desaturase | STE | ER |
| c88645_g1_i1 | 53 | 2.05 | fatty acid desaturase 4 | FAD4 | Pt |
| c94122_g2_i1 | 2097 | 33.65 | palmitoyl-monogalactosyldiacylglycerol delta-7 desaturase | FAD5 | Pt |
| c98080_g1_i5 | 538 | 8.42 | omega-6 fatty acid desaturase | FAD6 | Pt |
| c91818_g1_i1 | 54958 | 1369.99 | stearoyl-[acyl-carrier-protein] 9-desaturase | SAD6 | Pt |
| Fatty acid elongation | |||||
| c87821_g1_i1 | 53726 | 865.40 | beta-ketoacyl-CoA synthase | KCS | ER |
| c78850_g1_i1 | 14207 | 402.24 | very-long-chain 3-oxoacyl-CoA reductase 1 | KCR | ER |
| c140576_g1_i1 | 2 | 0.27 | very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | HCD | ER |
| c90092_g1_i1 | 4639 | 76.33 | very-long-chain enoyl-CoA reductase | ECR | ER |
| Fatty alcohol biosynthesis and oxidation | |||||
| c99851_g1_i2 | 69378 | 1613.05 | alcohol-forming fatty acyl-CoA reductase | FAR | ER |
| c95694_g1_i1 | 2554 | 39.02 | long-chain-alcohol oxidase FAO1 | FAO | ER |
| Triglyceride biosynthesis | |||||
| c92718_g1_i1 | 4782 | 60.72 | glycerol-3-phosphate dehydrogenase | G3PDH | ER |
| c93714_g1_i3 | 696 | 13.59 | glycerol-3-phosphate acyltransferase | GPAT | ER |
| c96688_g1_i1 | 6533 | 125.06 | 1-acylglycerol-3-phosphate O-acyltransferase | LPAAT | ER |
| c103123_g1_i1 | 2943 | 24.62 | phosphatidate phosphatase | PAP | ER |
| c102225_g2_i1 | 1510 | 16.53 | diacylglycerol O-acyltransferase | DAGT | ER |
| c93304_g1_i3 | 489 | 15.34 | monoacylglycerol lipase | MAGL | ER |
| c96753_g3_i1 | 1050 | 10.15 | Mono-/di-acylglycerol lipase | DAGL | ER |
| c103208_g4_i1 | 2523 | 32.09 | triacylglycerol lipase 2 | SDP2, TAGL | ER |
| Phospholipid metabolism | |||||
| c70471_g1_i1 | 1021 | 24.10 | CDP-diacylglycerol--inositol 3-phosphatidyltransferase 1 | CDIPT | Golgi |
| c102453_g1_i4 | 1210 | 7.99 | CDP-diacylglycerol--serine O-phosphatidyltransferase 1 | PSS | ER |
| c104411_g1_i1 | 147 | 6.60 | N-acylphosphatidylethanolamine synthase | PES | ER |
| c89493_g2_i1 | 325 | 11.93 | phosphatidylcholine:diacylglycerol cholinephosphotransferase 1 | PDCT | ER |
| c99283_g3_i1 | 1134 | 20.11 | phosphatidate cytidylyltransferase 1 | CTP (CDS) | ER |
| c95703_g2_i1 | 161 | 3.80 | phospholipase A1 | PLA1 | ER |
| c89578_g1_i1 | 3262 | 159.08 | phospholipase A2 | PLA2 | ER |
| c99134_g1_i1 | 2344 | 35.90 | phospholipase C | PLC | PM |
| c91314_g1_i1 | 9735 | 127.73 | phospholipase D | PLD | |
| c104602_g2_i3 | 1426 | 13.24 | phospholipase SGR2 | PLSGR2 | |
| c94956_g1_i2 | 2784 | 42.11 | lysophospholipase BODYGUARD 3 | BDG3 | |
| Wax ester biosynthesis | |||||
| c94816_g1_i1 | 3440 | 101.99 | long-chain-alcohol O-fatty-acyltransferase | WS, Jojoba | ER |
| c104303_g2_i3 | 1158 | 32.29 | O-acyltransferase WSD1 | WS, WSD1 | ER |
| Lipid transfer and storage | |||||
| c90684_g1_i1 | 173325 | 9748.18 | non-specific lipid-transfer protein | nsLTPs | Exc |
| c24977_g1_i1 | 38455 | 5214.36 | lipid-transfer protein DIR1 | LTP | Exc |
| c92005_g1_i2 | 96918 | 3779.71 | oleosin 18.2 kDa | OL | Cyt |
| c86220_g1_i1 | 30925 | 1036.72 | acyl carrier protein | ACP | Pt |
| c55316_g1_i1 | 34307 | 2454.44 | Acyl-CoA-binding protein | ACBP | Cyt |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, S.S.; Elseehy, M.M.; Aljuaid, B.S.; El-Shehawi, A.M. Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis. Plants 2020, 9, 588. https://doi.org/10.3390/plants9050588
Alotaibi SS, Elseehy MM, Aljuaid BS, El-Shehawi AM. Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis. Plants. 2020; 9(5):588. https://doi.org/10.3390/plants9050588
Chicago/Turabian StyleAlotaibi, Saqer S., Mona M. Elseehy, Bandar S. Aljuaid, and Ahmed M. El-Shehawi. 2020. "Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis" Plants 9, no. 5: 588. https://doi.org/10.3390/plants9050588
APA StyleAlotaibi, S. S., Elseehy, M. M., Aljuaid, B. S., & El-Shehawi, A. M. (2020). Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis. Plants, 9(5), 588. https://doi.org/10.3390/plants9050588





