Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life
Abstract
1. Overview
2. Embryogenesis, Seed Maturation, and Germination Rely on Mitochondria Functions and Cellular Metabolism
2.1. Seed as a Major Adaptation of Plants to Life on Land
2.2. Seed Quiescence, Dormancy, and Germination
3. Land Plant Mitochondria Genome Structures and Gene Expression, and Their Essential Roles in Successful Germination and Early Seedling Establishment
3.1. Mitochondria Biogenesis and Respiratory Reactivation during Seed Germination
3.2. Organization of Angiosperm Mitochondrial Genomes
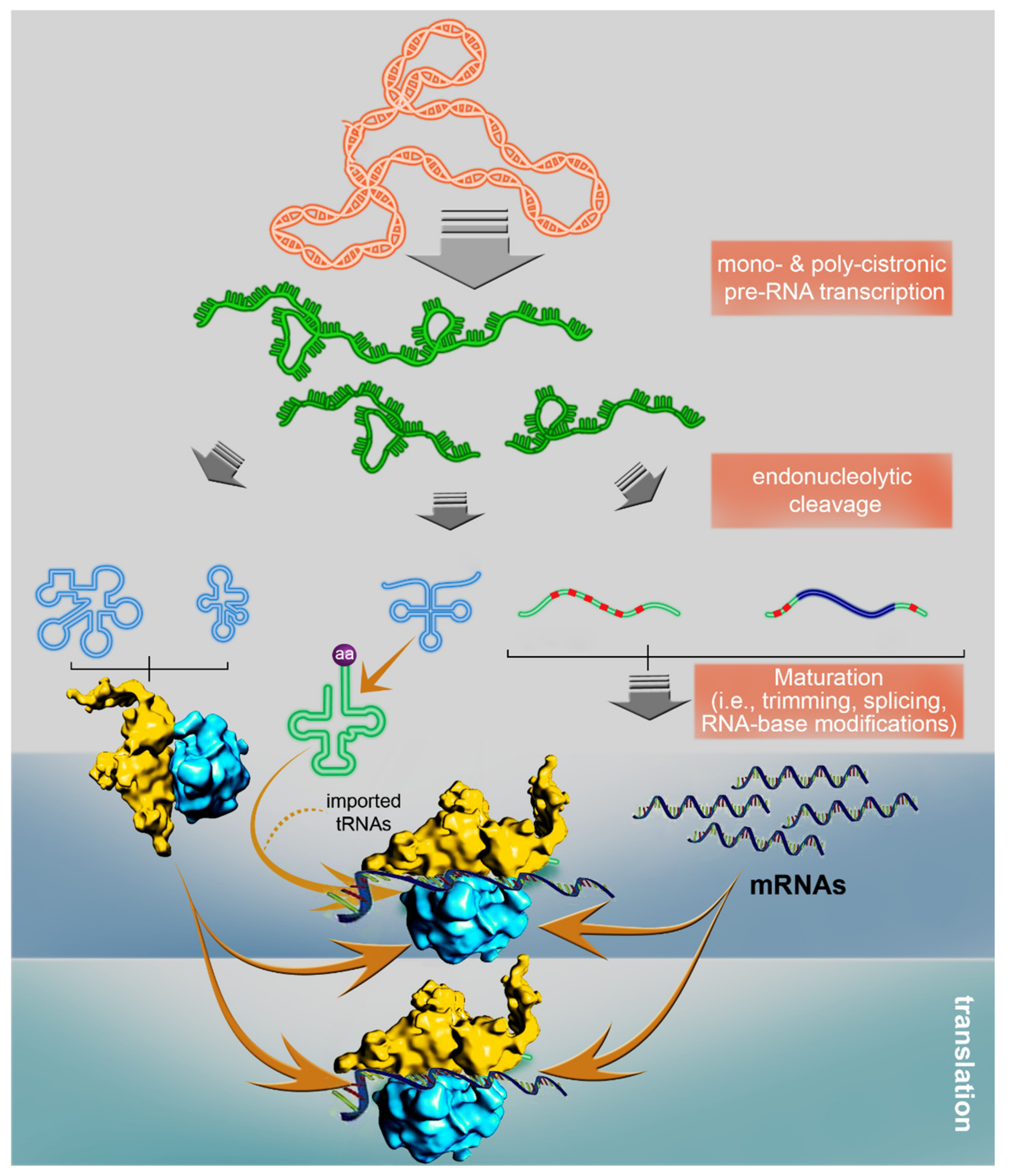
3.3. Plant Mitochondria Gene Expression, and Its Regulation during Early Germination Stages
3.3.1. The Biogenesis of the Import Machinery: A Molecular Switch for RNA Metabolism and Mitochondria Biogenesis during Early Germination
3.3.2. RNA Editing Plays a Key Role in the Regulation of mtDNA Expression
3.3.3. Presence and Putative Role of Group II-Type Introns in Angiosperm Mitochondria
3.3.4. A Mitochondrial Failure in Angiosperms Often Results in Altered Embryogenesis, Reduced Germination, and Retarded Seedlings Growth and Development
3.3.5. Angiosperm Mitochondria Gene Copy Numbers
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The timescale of early land plant evolution. Proc. Nalt. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef]
- Kenrick, P.; Crane, P.R. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.-F.; Lindquist, E.A.; Kamisugi, Y.; et al. The physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Cherns, L.; Raven, J.A. Could land-based early photosynthesizing ecosystems have bioengineered the planet in mid-Palaeozoic times? Palaeontology 2015, 58, 803–837. [Google Scholar] [CrossRef]
- Wellman, C.H.; Strother, P.K. The terrestrial biota prior to the origin of land plants (embryophytes): A review of the evidence. Palaeontology 2015, 58, 601–627. [Google Scholar] [CrossRef]
- Hermannus, P.; Sherard, W. Paradisus Batavus, Continens Plus Centum Plantas Affabrè ære Incisas & Descriptionibus Illustratas: Cui Accessit Catalogus Plantarum, Quas pro Tomus Nondum Editis, Delineandas; Elsevier: Leiden, The Netherlands, 1968. [Google Scholar]
- Nonogaki, H. Seed germination-The biochemical and molecular mechanisms. Breed. Sci. 2006, 56, 93–105. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. Book 2008, 6, e0119. [Google Scholar] [CrossRef]
- Ratajczak, E.; Małecka, A.; Ciereszko, I.; Staszak, A.M. Mitochondria are important determinants of the aging of seeds. Int. J. Mol. Sci. 2019, 20, 1568. [Google Scholar] [CrossRef]
- Czarna, M.; Kolodziejczak, M.; Janska, H. Mitochondrial proteome studies in seeds during germination. Proteomes 2016, 4, 19. [Google Scholar] [CrossRef]
- Law, S.R.; Narsai, R.; Whelan, J. Mitochondrial biogenesis in plants during seed germination. Mitochondrion 2014, 19, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Zmudjak, M.; Ostersetzer-Biran, O. RNA metabolism and transcript regulation. In Annual Plant Reviews; Logan, D.C., Ed.; Wiley: Oxford, UK, 2017; pp. 261–309. [Google Scholar] [CrossRef]
- Paszkiewicz, G.; Gualberto, J.M.; Benamar, A.; Macherel, D.; Logan, D.C. Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell 2017, 29, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.C.; Millar, A.H.; Sweetlove, L.J.; Hill, S.A.; Leaver, C.J. Mitochondrial biogenesis during germination in maize embryos. Plant Physiol. 2001, 125, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Kitazaki, K.; Kubo, T. Cost of having the largest mitochondrial genome: Evolutionary mechanism of plant mitochondrial genome. J. Botany 2010, 2010. [Google Scholar] [CrossRef]
- Fowler, S.; Roush, R.; Wise, J. Concepts of Biology: Early Plant Life; OpenStax: Houston, TX, USA, 2013. [Google Scholar]
- Wu, S.-Q. Land plants. In The Jehol Fossils; Chang, M.-M., Ed.; Academic Press: San Diego, CA, USA, 2008; pp. 166–177. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Nalt. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Adhikari, P.B.; Liu, X.; Wu, X.; Zhu, S.; Kasahara, R.D. Fertilization in flowering plants: An odyssey of sperm cell delivery. Plant Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Boavida, L.; McCormick, S. Gametophyte and Sporophyte. In eLS; Wiley: Hoboken, NJ, USA, 2010. [Google Scholar] [CrossRef]
- Boavida, L.C.; Becker, J.D.; Feijó, J.A. The making of gametes in higher plants. Int. J. Plant Dev. Biol. 2005, 49, 595–614. [Google Scholar] [CrossRef]
- Osorio, S.; Scossa, F.; Fernie, A. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Nascimento, V.L.; Medeiros, D.B.; Nunes-Nesi, A.; Ribeiro, D.M.; Zsögön, A.; Araújo, W.L. Modifications in organic acid profiles during fruit development and ripening: Correlation or causation? Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Seymour, G.B.; Ostergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef]
- Azzi, L.; Deluche, C.; Gévaudant, F.; Frangne, N.; Delmas, F.; Hernould, M.; Chevalier, C. Fruit growth-related genes in tomato. J. Exp. Bot. 2015, 66, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; de Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.S.; Elozeiri, A.A. Metabolic processes during seed germination. Adv. Seed Biol. 2017, 141–166. [Google Scholar]
- Kifle, T. Biology of seed development and germination physiology. Adv. Plants Agric. Res. 2018, 8. [Google Scholar] [CrossRef]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef]
- Harada, J.J. Seed maturation and control of germination. In Cellular and Molecular Biology of Plant Seed Development; Larkins, B.A., Vasil, I.K., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 1997; pp. 545–592. [Google Scholar] [CrossRef]
- Mayer, A.M.; Poljakoff-Mayber, A. The Germination of Seeds; Elsevier: Oxford, UK, 1982. [Google Scholar]
- Lubzens, E.; Cerda, J.; Clark, M. Dormancy and Resistance in Harsh Environments; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Née, G.; Xiang, Y.; Soppe, W.J.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef]
- Kwasniak-Owczarek, M.; Janska, H. In organello protein synthesis. Bio Protoc. 2014, 4, e1157. [Google Scholar] [CrossRef]
- Firenzuoli, A.; Vanni, P.; Ramponi, G.; Baccari, V. Changes in enzyme levels during germination of seeds of Triticum durum. Plant Physiol. 1968, 43, 260–264. [Google Scholar] [CrossRef]
- Bewley, J.D.; Bradford, K.; Hilhorst, H. Seeds: Physiology of Development, Germination and Dormancy; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Turner, J.F.; Turner, D.H. The regulation of carbohydrate metabolism. Annu. Rev. Plant Physiol. 1975, 26, 159–186. [Google Scholar] [CrossRef]
- Shen-Miller, J.; Mudgett, M.B.; Schopf, J.W.; Clarke, S.; Berger, R. Exceptional seed longevity and robust growth: Ancient Sacred Lotus from China. Am. J. Bot. 1995, 82, 1367–1380. [Google Scholar] [CrossRef]
- Sallon, S.; Solowey, E.; Cohen, Y.; Korchinsky, R.; Egli, M.; Woodhatch, I.; Simchoni, O.; Kislev, M. Germination, genetics, and growth of an ancient date seed. Science 2008, 320, 1464. [Google Scholar] [CrossRef] [PubMed]
- Yashina, S.; Gubin, S.; Maksimovich, S.; Yashina, A.; Gakhova, E.; Gilichinsky, D. Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost. Proc. Nalt. Acad. Sci. USA 2012, 109, 4008–4013. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and regulation of mitochondrial respiration in plants. Ann. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef]
- Schertl, P.; Braun, H.P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5, 163. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef]
- Li-Pook-Than, J.; Carrillo, C.; Bonen, L. Variation in mitochondrial transcript profiles of protein-coding genes during early germination and seedling development in wheat. Curr. Genet. 2004, V46, 374. [Google Scholar] [CrossRef]
- Howell, K.A.; Millar, A.H.; Whelan, J. Ordered assembly of mitochondria during rice germination begins with promitochondrial structures rich in components of the protein import apparatus. Plant Mol. Biol. 2006, 60, 201–223. [Google Scholar] [CrossRef]
- Narsai, R.; Law, S.R.; Carrie, C.; Xu, L.; Whelan, J. In depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis thaliana. Plant Physiol. 2011, 157, 1342–1362. [Google Scholar] [CrossRef]
- Law, S.R.; Narsai, R.; Taylor, N.L.; Delannoy, E.; Carrie, C.; Giraud, E.; Millar, A.H.; Small, I.; Whelan, J. Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 2012, 158, 1610–1627. [Google Scholar] [CrossRef]
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial cristae: Where beauty meets functionality. Trends Biochem. Sci. 2016, 41, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Benamar, A.; Tallon, C.; Macherel, D. Membrane integrity and oxidative properties of mitochondria isolated from imbibing pea seeds after priming or accelerated ageing. Seed Sci. Res. 2007, 13, 35–45. [Google Scholar] [CrossRef]
- Sultan, L.D.; Mileshina, D.; Grewe, F.; Rolle, K.; Abudraham, S.; Głodowicz, P.; Khan Niazi, A.; keren, I.; Shevtsov, S.; Klipcan, L.; et al. The reverse-transcriptase/RNA-maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. Plant Cell 2016, 28, 2805–2829. [Google Scholar] [CrossRef] [PubMed]
- Schimper, A.F.W. Uber die entwicklung der chlorophyllkorner und farbkorper. Bot. Zeit. 1883, 41, 105–112. [Google Scholar]
- Mereschkowsky, C. Uber natur und ursprung der chromatophoren im pflanzenreiche. Biol. Cent. 1905, 25, 293–604. [Google Scholar]
- Martin, W.; Mentel, M. The origin of mitochondria. Nat. Educ. 2010, 3, 58. [Google Scholar]
- Baur, E. Das Wesen und die Erblichkeitsverhältnisse der “Varietates albomarginatae hort” von Pelargonium zonale. Z Indukt Abstamm Vererb. 1909, 1, 330–351. [Google Scholar]
- Correns, C. Vererbungsversuche mit blass(gelb)grünen und buntblättrigen Sippen bei Mirabilis jalapa, Urtica pilulifera und Lunaria annua. Z Indukt Abstamm Vererb. 1909, 1, 291–329. [Google Scholar]
- Hagemann, R. The foundation of extranuclear inheritance: Plastid and mitochondrial genetics. Mol. Genet. Genom. 2010, 283, 199–209. [Google Scholar] [CrossRef]
- Ephrussi, B.H.H.; Tavlitzki, J. Action de L’acriflavine sur les Levures: La Mutation Petite Colonie; Masson et Cie: Paris, France, 1949. [Google Scholar]
- Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 1967, 14, IN225–IN226. [Google Scholar] [CrossRef]
- Gray, M.W. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 1999, 9, 678–687. [Google Scholar] [CrossRef]
- Ward, B.L.; Anderson, R.S.; Bendich, A.J. The mitochondrial genome is large and variable in a family of plants (Cucurbitaceae). Cell 1981, 25, 793–803. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Ann. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Bonen, L. Mitochondrial Genomes in Land Plants. In Molecular Life Sciences: An Encyclopedic Reference; Wells, R.D., Bond, J.S., Klinman, J., Masters, B.S.S., Eds.; Springer: New York, NY, USA, 2018; pp. 734–742. [Google Scholar] [CrossRef]
- The Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Sloan, D.; Alverson, A.; Chuckalovcak, J.; Wu, M.; McCauley, D.; Palmer, J.; Taylor, D. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; dePamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Cho, Y.; Mower, J.P.; Alverson, A.J.; Palmer, J.D. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol. Biol. Evol. 2008, 25, 1762–1777. [Google Scholar] [CrossRef]
- Andersson, S.G.E.; Zomorodipour, A.; Andersson, J.O.; Sicheritz-Pontén, T.; Alsmark, U.C.M.; Podowski, R.M.; Näslund, A.K.; Eriksson, A.-S.; Winkler, H.H.; Kurland, C.G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998, 396, 133–140. [Google Scholar] [CrossRef]
- Turmel, M.; Otis, C.; Lemieux, C. The mitochondrial genome of Chara vulgaris: Insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell 2003, 15, 1888–1903. [Google Scholar] [CrossRef]
- Small, I. Mitochondrial genomes as living ‘fossils’. BMC Biol. 2013, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. Plant mitochondrial genome diversity: The genomics revolution. In Plant Genome Diversity Volume 1: Plant Genomes, their Residents, and their Evolutionary Dynamics; Wendel, J.F., Greilhuber, J., Dolezel, J., Leitch, I.J., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 123–144. [Google Scholar]
- Guo, W.; Grewe, F.; Fan, W.; Young, G.J.; Knoop, V.; Palmer, J.D.; Mower, J.P. Ginkgo and Welwitschia Mitogenomes Reveal Extreme Contrasts in Gymnosperm Mitochondrial Evolution. Mol. Biol. Evol. 2016, 33, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylova, Y.V.; Terent’eva, L.Y. Huge mitochondrial genomes in embryophyta plants. Biol. Bull. Rev. 2017, 7, 497–505. [Google Scholar] [CrossRef]
- Gandini, C.L.; Sanchez-Puerta, M.V. Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Sci. Rep. 2017, 7, 43402. [Google Scholar] [CrossRef] [PubMed]
- Bendich, A.J. Reaching for the ring: The study of mitochondrial genome structure. Curr. Genet. 1993, 24, 279–290. [Google Scholar] [CrossRef]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef]
- Touzet, P.H.; Meyer, E. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 2014, 19, 166–171. [Google Scholar] [CrossRef]
- Shaya, F.; Gaiduk, S.; Keren, I.; Shevtsov, S.; Zemah, H.; Belausov, E.; Evenor, D.; Reuveni, M.; Ostersetzer-Biran, O. Expression of mitochondrial gene fragments within the tapetum induce male sterility by limiting the biogenesis of the respiratory machinery in transgenic tobacco. J. Integr. Plant Biol. 2012, 54, 115–130. [Google Scholar] [CrossRef]
- Villarreal, A.J.C.; Turmel, M.; Bourgouin-Couture, M.; Laroche, J.; Salazar Allen, N.; Li, F.-W.; Cheng, S.; Renzaglia, K.; Lemieux, C. Genome-wide organellar analyses from the hornwort Leiosporoceros dussii show low frequency of RNA editing. PLoS ONE 2018, 13, e0200491. [Google Scholar] [CrossRef]
- Xue, J.Y.; Liu, Y.; Li, L.; Wang, B.; Qiu, Y.L. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: Retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr. Genet. 2010, 56, 53–61. [Google Scholar] [CrossRef]
- Hecht, J.; Grewe, F.; Knoop, V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 2011, 3, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.; Cuenca, A.; Moller, I.M.; Seberg, O. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Maclean, A.E.; Hertle, A.P.; Ligas, J.; Bock, R.; Balk, J.; Meyer, E.H. Absence of complex I is associated with diminished respiratory chain function in European mistletoe. Curr. Biol. 2018, 28, 1614–1619.e1613. [Google Scholar] [CrossRef] [PubMed]
- Senkler, J.; Rugen, N.; Eubel, H.; Hegermann, J.; Braun, H.-P. Absence of complex I implicates rearrangement of the respiratory chain in European mistletoe. Curr. Biol. 2018, 28, 1606–1613.e1604. [Google Scholar] [CrossRef]
- John, U.; Lu, Y.; Wohlrab, S.; Groth, M.; Janouškovec, J.; Kohli, G.S.; Mark, F.C.; Bickmeyer, U.; Farhat, S.; Felder, M.; et al. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 2019, 5, eaav1110. [Google Scholar] [CrossRef]
- Yahalomi, D.; Atkinson, S.D.; Neuhof, M.; Chang, E.S.; Philippe, H.; Cartwright, P.; Bartholomew, J.L.; Huchon, D. A cnidarian parasite of salmon (Myxozoa Henneguya) lacks a mitochondrial genome. Proc. Nalt. Acad. Sci. USA 2020, 201909907. [Google Scholar] [CrossRef]
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Nalt. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef]
- Johnston, I.G.; Williams, B.P. Evolutionary Inference across Eukaryotes Identifies Specific Pressures Favoring Mitochondrial Gene Retention. Cell Syst. 2016, 2, 101–111. [Google Scholar] [CrossRef]
- Ostersetzer-Biran, O. Respiratory complex I and embryo development. J. Exp. Bot. 2016, 67, 1205–1207. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef]
- Schwarzländer, M.; König, A.-C.; Sweetlove, L.J.; Finkemeier, I. The impact of impaired mitochondrial function on retrograde signalling: A meta-analysis of transcriptomic responses. J. Exp. Bot. 2012, 63, 1735–1750. [Google Scholar] [CrossRef] [PubMed]
- Bonen, L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 2008, 8, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.G.; Colas des Francs-Small, C.; Ostersetzer-Biran, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5, 35. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Lampe, M.-K.; Sultan, L.D.; Ostersetzer-Biran, O. Organellar maturases: A window into the evolution of the spliceosome. BBA Bioenerg. 2015, 1847, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Waltz, F.; Nguyen, T.-T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rudinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsasser, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kuhn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2020, 101, 420–441. [Google Scholar] [CrossRef]
- Huang, S.; Shingaki-Wells, R.N.; Taylor, N.L.; Millar, A.H. The rice mitochondria proteome and its response during development and to the environment. Front. Plant Sci. 2013, 4, 16. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Ann. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Kolli, R.; Soll, J.; Carrie, C. Plant mitochondrial inner membrane protein insertion. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Giege, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Lamattina, L.; Bonnard, G.; Weil, J.H.; Grienenberger, J.M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef] [PubMed]
- Hiesel, R.; Wissinger, B.; Schuster, W.; Brennicke, A. RNA editing in plant mitochondria. Science 1989, 246, 1632–1634. [Google Scholar] [CrossRef]
- Freyer, R.; Kiefer-Meyer, M.C.; Kossel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Nalt. Acad. Sci. USA 1997, 94, 6285–6290. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2016, 8, 5–11. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol. Life Sci. 2011, 68, 567–586. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, K.; Zhang, D.; Wan, Y.; Wen, Y.; Lu, Q.; Wang, L. High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS ONE 2017, 12, e0185612. [Google Scholar] [CrossRef] [PubMed]
- Murik, O.; Chandran, S.; Nevo-Dinur, K.; Sultan, L.; Best, C.; Stein, Y.; Hazan, C.; Ostersetzer-Biran, O. The topologies of N6-Adenosine methylation (m6A) in land plant mitochondria and their putative effects on organellar gene-expression. Plant J. 2019, 101, 1269–1286. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Lopes Dos Santos, A.; Otis, C.; Sergerie, R.; Lemieux, C. Tracing the evolution of the plastome and mitogenome in the Chloropicophyceae uncovered convergent tRNA gene losses and a variant plastid genetic code. Genome Biol. Evol. 2019, 11, 1275–1292. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Kusumegi, T.; Tsudzuki, T.; Sugiura, M. RNA editing sites in tobacco chloroplast transcripts: Editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet. 1999, 262, 462–467. [Google Scholar] [CrossRef]
- Karcher, D.; Bock, R. The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J. Biol. Chem. 2002, 277, 5570–5574. [Google Scholar] [CrossRef]
- Bock, R.; Hagemann, R.; Kossel, H.; Kudla, J. Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids--a new regulatory mechanism? Mol. Gen. Genet. 1993, 240, 238–244. [Google Scholar] [CrossRef]
- Hirose, T.; Fan, H.; Suzuki, J.Y.; Wakasugi, T.; Tsudzuki, T.; Kossel, H.; Sugiura, M. Occurrence of silent RNA editing in chloroplasts: Its species specificity and the influence of environmental and developmental conditions. Plant Mol. Biol. 1996, 30, 667–672. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.L.; Hanson, M.R. Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res. 2003, 31, 2586–2594. [Google Scholar] [CrossRef]
- Guillaumot, D.; Lopez-Obando, M.; Baudry, K.; Avon, A.; Rigaill, G.; Falcon de Longevialle, A.; Broche, B.; Takenaka, M.; Berthomé, R.; De Jaeger, G.; et al. Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Nalt. Acad. Sci. USA 2017, 114, 8877–8882. [Google Scholar] [CrossRef]
- Dahan, J.; Tcherkez, G.; Macherel, D.; Benamar, A.; Belcram, K.; Quadrado, M.; Arnal, N.; Mireau, H. Disruption of the CYTOCHROME C OXIDASE DEFICIENT1 gene leads to cytochrome c oxidase depletion and reorchestrated respiratory metabolism in Arabidopsis. Plant Physiol. 2014, 166, 1788–1802. [Google Scholar] [CrossRef]
- Small, I.D.; Peeters, N. The PPR motif-A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 45–47. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bonen, L.; Vogel, J. The ins and outs of group II introns. Trends Genet. 2001, 17, 322–331. [Google Scholar] [CrossRef]
- Zmudjak, M.; Ostersetzer-Biran, O. RNA metabolism and transcript regulation. Annu. Plant Rev. Online 2018, 143–184. [Google Scholar] [CrossRef]
- Robart, A.R.; Zimmerly, S. Group II intron retroelements: Function and diversity. Cytogenet. Genome Res. 2005, 110, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Lambowitz, A.M.; Zimmerly, S. Group II introns: Mobile ribozymes that invade DNA. Perspect. Biol. 2011, 3, 1–19. [Google Scholar] [CrossRef]
- Zimmerly, S.; Semper, C. Evolution of group II introns. Mobile DNA 2015, 6, 1–19. [Google Scholar] [CrossRef]
- Palmer, J.D.; Adams, K.L.; Cho, Y.; Parkinson, C.L.; Qiu, Y.L.; Song, K. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Nalt. Acad. Sci. USA 2000, 97, 6960–6966. [Google Scholar] [CrossRef]
- Guo, W.; Mower, J.P. Evolution of plant mitochondrial intron-encoded maturases: Frequent lineage-specific loss and recurrent intracellular transfer to the nucleus. J. Mol. Evol. 2013, 77, 43–54. [Google Scholar] [CrossRef]
- Becker, B.; Marin, B. Streptophyte algae and the origin of embryophytes. Ann. Bot. 2009, 103, 999–1004. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Xu, T.; Zucchi, P.; Bogorad, L. Subpopulations of chloroplast ribosomes change during photoregulated development of Zea mays leaves: Ribosomal proteins L2, L21, and L29. Proc. Nalt. Acad. Sci. USA 1999, 96, 8997–9002. [Google Scholar] [CrossRef] [PubMed]
- Dalby, S.J.; Bonen, L. Impact of low temperature on splicing of atypical group II introns in wheat mitochondria. Mitochondrion 2013, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Small, I. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 2014, 100, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Fromm, S.; Braun, H.P.; Peterhansel, C. Mitochondrial gamma carbonic anhydrases are required for complex I assembly and plant reproductive development. New Phytol. 2016, 211, 194–207. [Google Scholar] [CrossRef]
- Fromm, S.; Going, J.; Lorenz, C.; Peterhansel, C.; Braun, H.P. Depletion of the “gamma-type carbonic anhydrase-like” subunits of complex I affects central mitochondrial metabolism in Arabidopsis thaliana. Biochim. Biophys. Acta 2016, 1857, 60–71. [Google Scholar] [CrossRef]
- Fromm, S.; Senkler, J.; Eubel, H.; Peterhansel, C.; Braun, H.P. Life without complex I: Proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J. Exp. Bot. 2016, 67, 3079–3093. [Google Scholar] [CrossRef]
- Keren, I.; Bezawork-Geleta, A.; Kolton, M.; Maayan, I.; Belausov, E.; Levy, M.; Mett, A.; Gidoni, D.; Shaya, F.; Ostersetzer-Biran, O. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 2009, 15, 2299–2311. [Google Scholar] [CrossRef]
- Colas des Francs-Small, C.; Kroeger, T.; Zmudjak, M.; Ostersetzer-Biran, O.; Rahimi, N.; Small, I.; Barkan, A. A PORR domain protein required for rpl2 and ccmFc intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012, 69, 996–1005. [Google Scholar] [CrossRef]
- Keren, I.; Tal, L.; Colas des Francs-Small, C.; Araújo, W.L.; Shevtsov, S.; Shaya, F.; Fernie, A.R.; Small, I.; Ostersetzer-Biran, O. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 2012, 71, 413–426. [Google Scholar] [CrossRef]
- Zmudjak, M.; Colas des Francs-Small, C.; Keren, I.; Shaya, F.; Belausov, E.; Small, I.; Ostersetzer-Biran, O. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 2013, 199, 379–394. [Google Scholar] [CrossRef]
- Cohen, S.; Zmudjak, M.; Colas des Francs-Small, C.; Malik, S.; Shaya, F.; Keren, I.; Belausov, E.; Many, Y.; Brown, G.G.; Small, I.; et al. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 2014, 78, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Knoop, V. Seed plant mitochondrial genomes: Complexity evolving. In Genomics of chloroplasts and mitochondria; Bock, R., Knoop, V., Eds.; Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2012; pp. 175–200. [Google Scholar]
- Wang, D.-Y.; Zhang, Q.; Liu, Y.; Lin, Z.-F.; Zhang, S.-X.; Sun, M.-X.; Sodmergen. The levels of male gametic mitochondrial DNA are highly regulated in angiosperms with regard to mitochondrial inheritance. Plant Cell 2010, 22, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Bendich, A.J.; Gauriloff, L.P. Morphometric analysis of cucurbit mitochondria: The relationship between chondriome volume and DNA content. Protoplasma 1984, 119, 1–7. [Google Scholar] [CrossRef]
- Arimura, S.-i.; Yamamoto, J.; Aida, G.P.; Nakazono, M.; Tsutsumi, N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Nalt. Acad. Sci. USA 2004, 101, 7805–7808. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.D.; Soltis, D.E.; Soltis, P.S. The age of the angiosperms: A molecular timescale without a clock. Evolution 2005, 59, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Magallón, S. Using fossils to break long branches in molecular dating: A comparison of relaxed clocks applied to the origin of angiosperms. Syst. Biol. 2010, 59, 384–399. [Google Scholar] [CrossRef]
- Planchard, N.; Bertin, P.; Quadrado, M.; Dargel-Graffin, C.; Hatin, I.; Namy, O.; Mireau, H. The translational landscape of Arabidopsis mitochondria. Nucleic Acids Res. 2018, 46, 6218–6228. [Google Scholar] [CrossRef]
| Phylogeny | Organism | mtDNA Size (Kb) | Accession No. | C→U Editing | Group I Introns*1 | Group II Introns | |
|---|---|---|---|---|---|---|---|
| Bacteria | Rickettsia*2 | Rickettsia proazekii | (1109.3) | NC_020993 | - | - | - |
| Plantae | Green Alga | Caulerpa ashmeadii | 197.4 | NC-045849 | - | - | - |
| Chara vulgaris | 67.7 | NC-005255 | - | 13 | 14 | ||
| Chlamydomonas reinhardtii | 15.8 | NC-001638 | - | - | - | ||
| Chloroparvula pacifica | 49.7 | NC-042603 | (?)*3 | 2 | - | ||
| Land Plants*4 | Arabidopsis thalianaA | 367.8 | NC-037304 | + | - | 23 | |
| Climacium americanumB | 105.1 | NC_024515 | + | 2 | 23 | ||
| Brassica oleracea (Cauliflower)A | 360.2 | NC-016118 | + | - | 23 | ||
| Ginkgo bilobaG | 346.5 | NC_027976 | + | - | 20 | ||
| Oryza sativa (Rice)A | 491.5 | NC_007886 | + | - | 25 | ||
| Silene latifoliaA | 253.4 | NC_014487 | + | - | 13 | ||
| Triticum aestivum (Wheat)A | 452.5 | NC-036024 | + | - | 23 | ||
| Welwitschia mirabilisG | 978.9 | NC_029130 | + | - | 17 | ||
| Zea mays (Corn)A | 569.6 | NC-007982 | + | - | 22 | ||
| Fungi | Pichia canadensis | 27.7 | NC-001762 | - | 2 | - | |
| Saccharomyces cerevisiae | 78.9 | NC-027264 | - | 3 | 9 | ||
| Schizosaccharomyces pombe | 19.4 | NC-001326 | - | 2 | 1 | ||
| Animalia | Insects | Anopheles gambiae (Mosquito) | 15.4 | NC-002084 | - | - | - |
| Apis mellifera (Honeybee) | 16.3 | NC-001566 | - | - | - | ||
| Drosophila melanogaster (Fruit Fly) | 19.5 | NC-024511 | - | - | - | ||
| Formica fusca (Black Ant) | 16.6 | NC-026132 | - | - | - | ||
| Chnidarians | Aurelia aurita | 16.9 | NC-008446 | - | - | - | |
| Chrysopathes formosa | 18.4 | NC-008411 | - | 1 | - | ||
| Hydra oligactis | 16.3 | NC-010214 | - | - | - | ||
| Metridium senile | 17.4 | NC-000933 | - | 2 | - | ||
| Mammals | Mus musculus (Mouse) | 16.3 | NC-005089 | - | - | - | |
| Pan troglodytes (Chimpanzee) | 16.6 | NC-001643 | - | - | - | ||
| Homo sapiens (Human) | 16.6 | NC-012920 | - | - | - | ||
| Homo sapiens neanderthalensis | 16.6 | NC-011137 | - | - | - | ||
| Protista | Dictyostelium discoideum | 55.6 | NC-000895 | - | 6 | - | |
| Monosiga brevicollis | 76.6 | NC-004309 | - | 4 | - | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Best, C.; Mizrahi, R.; Ostersetzer-Biran, O. Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life. Plants 2020, 9, 598. https://doi.org/10.3390/plants9050598
Best C, Mizrahi R, Ostersetzer-Biran O. Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life. Plants. 2020; 9(5):598. https://doi.org/10.3390/plants9050598
Chicago/Turabian StyleBest, Corinne, Ron Mizrahi, and Oren Ostersetzer-Biran. 2020. "Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life" Plants 9, no. 5: 598. https://doi.org/10.3390/plants9050598
APA StyleBest, C., Mizrahi, R., & Ostersetzer-Biran, O. (2020). Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life. Plants, 9(5), 598. https://doi.org/10.3390/plants9050598






