Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants
Abstract
:1. Introduction
2. Classes, Features, and Uses of Antiviral Art-sRNAi Tools
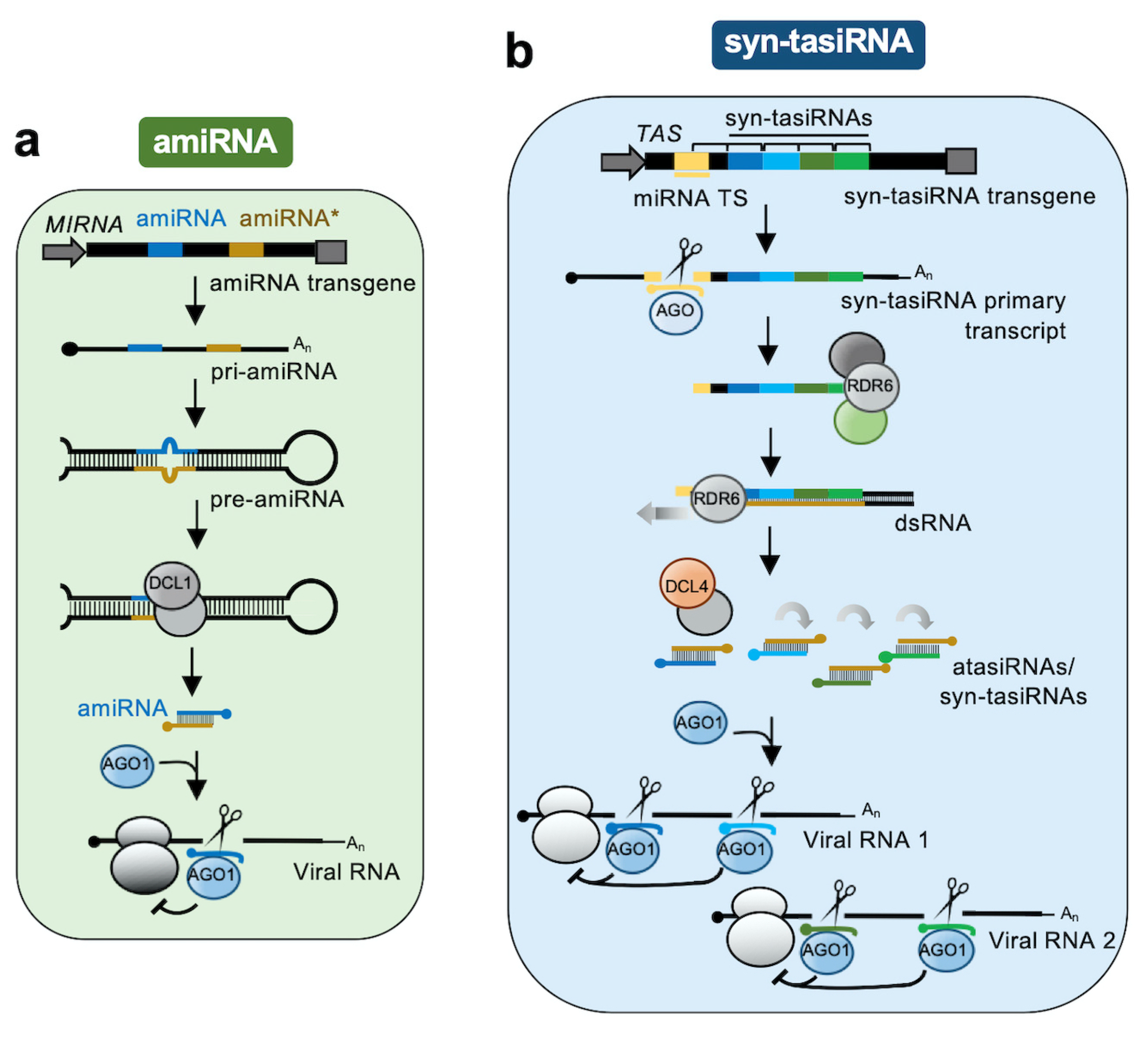
2.1. AmiRNAs
2.2. Syn-tasiRNAs
3. Design of Antiviral Art-sRNAs
3.1. General Design Rules
3.2. Selection of Target Sequences in Viral RNAs
4. Recent Advances in Art-sRNAi for Enhanced Antiviral Resistance
4.1. Identification of Effective Art-sRNAs with High Antiviral Activity
4.2. Co-Expression of Multiple Art-sRNAs for Viral RNA Multi-Targeting
4.3. Other
5. Application of Art-sRNAs to Control Viral Diseases in Crops
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AB | asymmetric bulge |
| art-sRNA | artificial small RNA |
| art-sRNAi | art-sRNA-based RNA interference |
| AGO | argonaute |
| amiRNA | artificial microRNA |
| atasiRNA | artificial trans-acting small interfering RNA |
| CP | coat protein |
| DCL | Dicer-like |
| dsRNA | double-stranded RNA |
| esiRNA | immunologically effective small interfering RNA |
| hp | hairpin |
| MIRNA | microRNA |
| pri-amiRNA | primary artificial microRNA precursor |
| pri-syn-tasiRNA | primary synthetic trans-acting small interfering RNA precursor |
| RdRp | RNA-dependent RNA polymerase |
| RNAi | RNA interference |
| sRNA | small RNA |
| syn-tasiRNA | synthetic trans-acting small interfering RNA |
| VSR | viral silencing suppressor protein |
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Gaffar, F.Y.; Koch, A. Catch Me If You Can! RNA Silencing-Based Improvement of Antiviral Plant Immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senthil-Kumar, M.; Mysore, K.S. Caveat of RNAi in Plants: The Off-Target Effect. In RNAi and Plant Gene Function Analysis: Methods and Protocols; Kodama, H., Komamine, A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 13–25. ISBN 978-1-61779-123-9. [Google Scholar]
- López-Dolz, L.; Spada, M.; Daròs, J.-A.; Carbonell, A. Fine-Tune Control of Targeted RNAi Efficacy by Plant Artificial Small RNAs. Nucleic Acids Res. 2020, in press. [Google Scholar] [CrossRef]
- Carbonell, A. Artificial small RNA-based strategies for effective and specific gene silencing in plants. In Plant Gene Silencing: Mechanisms and Applications; Dalmay, T., Ed.; CABI Publishing: Wallingford, Oxfordshire, UK, 2017; pp. 110–127. ISBN 978-1-78064-767-8. [Google Scholar]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. For. Cell Mol. Biol. 2008, 53, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Simon-Mateo, C.; Garcia, J.A. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.S.; Wu, H.W.; Elena, S.F.; Chen, K.C.; Niu, Q.W.; Yeh, S.D.; Chen, C.C.; Chua, N.H. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 2009, 5, e1000312. [Google Scholar] [CrossRef] [Green Version]
- Lafforgue, G.; Martinez, F.; Sardanyes, J.; de la Iglesia, F.; Niu, Q.W.; Lin, S.S.; Sole, R.V.; Chua, N.H.; Daros, J.A.; Elena, S.F. Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. J. Virol. 2011, 85, 9686–9695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonell, A.; Lisón, P.; Daròs, J.-A. Multi-targeting of viral RNAs with synthetic trans-acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.J. Artificial trans-acting small interfering RNA: A tool for plant biology study and crop improvements. Planta 2014, 239, 1139–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Daròs, J.-A. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, A.; Lopez, C.; Daròs, J.-A. Fast-Forward Identification of Highly Effective Artificial Small RNAs Against Different Tomato spotted wilt virus Isolates. Mol. Plant Microbe Interact. 2019, 32, 142–156. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, A. Secondary Small Interfering RNA-Based Silencing Tools in Plants: An Update. Front. Plant Sci. 2019, 10, 687. [Google Scholar] [CrossRef] [Green Version]
- Felippes, F.F.; Wang, J.W.; Weigel, D. MIGS: miRNA-induced gene silencing. Plant J. Cell Mol. Biol. 2012, 70, 541–547. [Google Scholar] [CrossRef]
- Zhao, M.; San Leon, D.; Mesel, F.; Garcia, J.A.; Simon-Mateo, C. Assorted Processing of Synthetic Trans-Acting siRNAs and Its Activity in Antiviral Resistance. PLoS ONE 2015, 10, e0132281. [Google Scholar] [CrossRef]
- Singh, A.; Taneja, J.; Dasgupta, I.; Mukherjee, S.K. Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol. Plant Pathol. 2015, 16, 724–734. [Google Scholar] [CrossRef]
- Singh, A.; Mohorianu, I.; Green, D.; Dalmay, T.; Dasgupta, I.; Mukherjee, S.K. Artificially induced phased siRNAs promote virus resistance in transgenic plants. Virology 2019, 537, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Hill, S.T.; Carrington, J.C.; Carbonell, A. P-SAMS: A web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 2016, 32, 157–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Hao, J.; Li, J.; Baker, B.; Luo, L. Artificial microRNA-mediated resistance to cucumber green mottle mosaic virus in Nicotiana benthamiana. Planta 2019, 250, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Wagaba, H.; Patil, B.L.; Mukasa, S.; Alicai, T.; Fauquet, C.M.; Taylor, N.J. Artificial microRNA-derived resistance to Cassava brown streak disease. J. Virol. Methods 2016, 231, 38–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.G.; Wang, C.H.; Fang, R.X.; Guo, H.S. Artificial MicroRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, H.; Zhang, J.; Zhang, C.; Gong, P.; Ziaf, K.; Xiao, F.; Ye, Z. Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res. 2011, 20, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef]
- Mitter, N.; Zhai, Y.; Bai, A.X.; Chua, K.; Eid, S.; Constantin, M.; Mitchell, R.; Pappu, H.R. Evaluation and identification of candidate genes for artificial microRNA-mediated resistance to tomato spotted wilt virus. Virus Res. 2016, 211, 151–158. [Google Scholar] [CrossRef]
- Lafforgue, G.; Martinez, F.; Niu, Q.W.; Chua, N.H.; Daros, J.A.; Elena, S.F. Improving the effectiveness of artificial microRNA (amiR)-mediated resistance against Turnip mosaic virus by combining two amiRs or by targeting highly conserved viral genomic regions. J. Virol. 2013, 87, 8254–8256. [Google Scholar] [CrossRef] [Green Version]
- Kung, Y.J.; Lin, S.S.; Huang, Y.L.; Chen, T.C.; Harish, S.S.; Chua, N.H.; Yeh, S.D. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol. Plant Pathol. 2012, 13, 303–317. [Google Scholar] [CrossRef]
- Ali, I.; Amin, I.; Briddon, R.W.; Mansoor, S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol. J. 2013, 10, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jelly, N.S.; Schellenbaum, P.; Walter, B.; Maillot, P. Transient expression of artificial microRNAs targeting Grapevine fanleaf virus and evidence for RNA silencing in grapevine somatic embryos. Transgenic Res. 2012, 21, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Z.; Han, Q.J.; Jiang, F.; Sun, R.Z.; Fan, Z.H.; Zhu, C.X.; Wen, F.J. Effects of the sequence characteristics of miRNAs on multi-viral resistance mediated by single amiRNAs in transgenic tobacco. Plant Physiol. Biochem. 2014, 77, 90–98. [Google Scholar] [CrossRef]
- Yu, R.; Chen, C.; Cao, W.; Liu, H.; Zhou, S.; Song, Y.; Zhu, C. High-degree and broad-spectrum resistance mediated by a combination of NIb siRNA and miRNA suppresses replication of necrotic and common strains of potato virus Y. Arch. Virol. 2018, 163, 3073–3081. [Google Scholar] [CrossRef]
- Vu, T.V.; Choudhury, N.R.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013, 172, 35–45. [Google Scholar] [CrossRef]
- Gago-Zachert, S.; Schuck, J.; Weinholdt, C.; Knoblich, M.; Pantaleo, V.; Grosse, I.; Gursinsky, T.; Behrens, S.-E. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro. Nucleic Acids Res. 2019, 47, 9343–9357. [Google Scholar] [CrossRef] [Green Version]
- Kis, A.; Tholt, G.; Ivanics, M.; Varallyay, E.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef]
- Xuan, N.; Zhao, C.; Peng, Z.; Chen, G.; Bian, F.; Lian, M.; Liu, G.; Wang, X.; Bi, Y. Development of transgenic maize with anti-rough dwarf virus artificial miRNA vector and their disease resistance. Chin. J. Biotechnol. 2015, 31, 1375–1386. [Google Scholar]
- Zhang, H.; Feng, H.; Lu, X.; Wang, C.; Yang, W.; Li, F. An asymmetric bulge enhances artificial microRNA-mediated virus resistance. Plant Biotechnol. J. 2019, 18, 608–610. [Google Scholar] [CrossRef] [Green Version]
- Petchthai, U.; Yee, C.S.L.; Wong, S.M. Resistance to CymMV and ORSV in artificial microRNA transgenic Nicotiana benthamiana plants. Sci. Rep. 2018, 8, 9958. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lin, C.; Du, J.; Song, Y.; Jiang, M.; Liu, H.; Zhou, S.; Wen, F.; Zhu, C. Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell Tiss Organ. Cult. 2016, 126, 1–13. [Google Scholar] [CrossRef]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2012, 10, 150–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravelonandro, M.; Scorza, R.; Briard, P. Innovative RNAi Strategies and Tactics to Tackle Plum Pox Virus (PPV) Genome in Prunus domestica-Plum. Plants 2019, 8, 565. [Google Scholar] [CrossRef] [Green Version]
- Csorba, T.; Kontra, L.; Burgyan, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, A.; Daròs, J.-A. Design, Synthesis, and Functional Analysis of Highly Specific Artificial Small RNAs with Antiviral Activity in Plants. Methods Mol. Biol. 2019, 2028, 231–246. [Google Scholar]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014, 165, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Li, J.F.; Zhang, D.; Sheen, J. Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat. Protoc. 2014, 9, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Li, J.F.; Chung, H.S.; Niu, Y.; Bush, J.; McCormack, M.; Sheen, J. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 2013, 25, 1507–1522. [Google Scholar] [CrossRef] [Green Version]
- Szittya, G.; Moxon, S.; Pantaleo, V.; Toth, G.; Rusholme Pilcher, R.L.; Moulton, V.; Burgyan, J.; Dalmay, T. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010, 6, e1000838. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Burgyan, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 3797–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miozzi, L.; Gambino, G.; Burgyan, J.; Pantaleo, V. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 2013, 14, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Schuck, J.; Gursinsky, T.; Pantaleo, V.; Burgyan, J.; Behrens, S.E. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 2013, 41, 5090–5103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargason, J.M.; Szittya, G.; Burgyan, J.; Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef] [Green Version]
- Cuperus, J.T.; Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Burke, R.T.; Takeda, A.; Sullivan, C.M.; Gilbert, S.D.; Montgomery, T.A.; Carrington, J.C. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010, 17, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Koenig, D.; Weigel, D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 2012, 109, 2461–2466. [Google Scholar] [CrossRef] [Green Version]
- Narjala, A.; Nair, A.; Tirumalai, V.; Hari Sundar, G.V.; Shivaprasad, P.V. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res. 2020, 48, 3103–3118. [Google Scholar] [CrossRef]
- Yang, M.; Woolfenden, H.C.; Zhang, Y.; Fang, X.; Liu, Q.; Vigh, M.L.; Cheema, J.; Yang, X.; Norris, M.; Yu, S.; et al. In vivo mRNA structure regulates miRNA cleavage in Arabidopsis | bioRxiv. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA Based Genetic Engineering for Plant Viral Resistance: Application in Crop Protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Tenllado, F.; Diaz-Ruiz, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonell, A.; Martinez de Alba, A.E.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Shimura, H.; Masuta, C. Advancing toward commercial application of RNA silencing-based strategies to protect plants from viral diseases. J. Gen. Plant Pathol. 2019, 85, 321–328. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55. [Google Scholar] [CrossRef]
- Tenllado, F.; Llave, C.; Diaz-Ruiz, J.R. RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res. 2004, 102, 85–96. [Google Scholar] [CrossRef]
- Daros, J.A.; Aragones, V.; Cordero, T. A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli. Sci. Rep. 2018, 8, 1904. [Google Scholar] [CrossRef]
- Yin, G.; Sun, Z.; Liu, N.; Zhang, L.; Song, Y.; Zhu, C.; Wen, F. Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl. Microbiol. Biotechnol. 2009, 84, 323–333. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef] [Green Version]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]


| Art-sRNA Class/Type of Precursor | Precursor(s) | No. Art-sRNAs | Target Virus(es) 1 | Target Region(s) 2 | Effects | Type of Expression 3/Transformation Method | Plant Species | Ref. |
|---|---|---|---|---|---|---|---|---|
| amiRNA/single monocistronic | AthMIR156 | 1 | CGMMV | CP | High protection. | TE/leaf agroinfiltration | Nicotiana benthamiana | [25] |
| AthMIR159a | 1 | CBSV | P1, P3, NIb, 3′UTR | High protection when targeting P1 or. NIb (also against UCBSV). | SE/leaf disc | Nicotiana benthamiana | [26] | |
| 1 | CMV | 3′ UTR (RNA3) | High protection when target site is not in tRNA-like structures. | SE/floral dip | Arabidopsis thaliana | [27] | ||
| 1 | CMV | 3′ UTR (RNA3) | High protection when target site is not in tRNA-like structures. | SE/leaf disc | Nicotiana tabacum | [27] | ||
| 1 | CMV | 2a/2b, 3′ UTR | Higher protection when targeting 2a/2b. | SE/cotyledonary explants | Solanum lycopersicum | [28] | ||
| 1 | PVX | p25 | High protection, even at increased viral pressure. | SE/leaf disc | Nicotiana tabacum | [29] | ||
| 1 | PVY | HC-Pro | High protection, even at increased viral pressure. | SE/leaf disc | Nicotiana tabacum | [29] | ||
| 1 | TSWV | N, NSs | High protection when targeting N. No protection when targeting NSs. | TE/leaf agroinfiltration | Nicotiana benthamiana | [30] | ||
| SE/leaf disc | Nicotiana tabacum | |||||||
| 1 | TuMV | HC-Pro | High protection. | SE/floral dip | Arabidopsis thaliana | [10] | ||
| Virus escapes emerge at subinhibitory amiRNA concentrations. | SE/floral dip | Arabidopsis thaliana | [13] | |||||
| Intermediate protection. | SE/floral dip | Arabidopsis thaliana | [31] | |||||
| 1 | TuMV | CP | High protection. | SE/floral dip | Arabidopsis thaliana | [31] | ||
| 1 | TYMV | P69 | High protection. | SE/floral dip | Arabidopsis thaliana | [10] | ||
| 1 | UCBSV | P1, P3, CI, NIb, CP, 3′UTR | High protection when targeting P1 or. CP (lower against UCBSV). | SE/leaf disc | Nicotiana benthamiana | [26] | ||
| 1 | WSMoV | A, B1, B2, C, D, E (RdRP) | Intermediate protection when targeting B2 and D. | SE/leaf disc | Nicotiana benthamiana | [32] | ||
| AthMIR164 | 1 | CGMMV | MP | High protection. | TE/leaf agroinfiltration | Nicotiana benthamiana | [25] | |
| AthMIR167b | 1 | PVX | p25 | Intermediate protection. Broken resistance after re-inoculation. | SE/leaf disc | Nicotiana tabacum | [29] | |
| 1 | PVY | HC-Pro | ||||||
| AthMIR169a | 1 | CLCuBuV | V2 | Low or high protection when the precursor was or was not modified, respectively. | SE/leaf disc | Nicotiana benthamiana | [33] | |
| AthMIR171a | 1 | CMV | 2b | Inhibition of 2b silencing suppressor function. | TE/leaf agroinfiltration | Nicotiana benthamiana, | [34] | |
| 63.3% of the lines were resistant. | SE/leaf disc | Nicotiana tabacum | ||||||
| 1 | CGMMV | Rep | High protection. | TE/leaf agroinfiltration | Nicotiana benthamiana | [25] | ||
| 1 | PVX | p25 | Intermediate protection. Broken resistance after re-inoculation. | SE/leaf disc | Nicotiana tabacum | [29] | ||
| 1 | PVY | HC-Pro | ||||||
| AthMIR319a | 1 | GFLV | CP | AmiRNAs are active against GFLV target sites located in a GUS mRNA sensor. | TE/somatic embryos at cotyledonary stage | Vitis vinifera | [35] | |
| 1 | PVY | CI, NIa, NIb, CP | Higher protection when targeting NIb or CP. | SE/leaf disc | Nicotiana tabacum | [36] | ||
| 1 | PVYO + PVYN | NIb (PVYO) + NIb (PVYN) | 33% and 17% of the lines were resistant to PVYO and PVYN, respectively. | SE/leaf disc | Nicotiana tabacum | [37] | ||
| 1 | TEV | CI, NIa, NIb, CP | Higher protection when targeting NIb or CP. | SE/leaf disc | Nicotiana tabacum | [36] | ||
| 1 | ToLCNDV | AV1, AV1 + AV2 | High tolerance when targeting AV1 + AV2. Moderate tolerance when targeting AV1. | SE/cotyledonary explants | Solanum lycopersicum | [38] | ||
| AthMIR390a | 1 | PSTVd | TL, C, V [PSTVd(+)], | Delay of viroid accumulation in all cases. | TE/leaf agroinfiltration | Nicotiana benthamiana | [17] | |
| 1 | PSTVd | TL, P, C, V, TR [PSTVd(−)] | Delay of viroid accumulation when targeting TL and C. | |||||
| 1 | TBSV | 5′ terminus TBSV(+) RNA | 40–90% of plants were symptom-free. | TE/leaf agroinfiltration | Nicotiana benthamiana | [39] | ||
| 1 | TSWV | N, NSm, NSs, RdRP | 50–100% of the plants did not accumulate TSWV when targeting NSm or RdRP. | TE/leaf agroinfiltration | Nicotiana benthamiana | [18] | ||
| 1 | TSWV | RdRP | 22% of the lines were resistant. | SE/cotyledonary explants | Solanum lycopersicum | [14] | ||
| HvuMIR171a | 1 | WDV | MP, Rep, RepA, RepA + Rep | AmiRNAs against Rep and RepA + Rep were selected based on a reporter system. | TE/leaf agroinfiltration | Nicotiana benthamiana | [40] | |
| SlyMIR159a | 1 | ToLCNDV | AV1, AV1 + AV2 | High tolerance when targeting AV1 + AV2. Moderate tolerance when targeting AV1. | SE/cotyledonary explants | Solanum lycopersicum | [38] | |
| SlyMIR168a | 1 | ToLCNDV | AV1, AV1 + AV2 | High tolerance when targeting AV1 + AV2. No accumulation of amiRNAs against AV1. | ||||
| ZmaMIR159a | 1 | RBSDV | P6 | High protection. | SE/ear immature embryos | Zea mays | [41] | |
| amiRNA/single monocistronic in tandem repeats | AthMIR159a | 2 | PVX + PVY | P25 (PVX) + HC-Pro (PVY) | High protection against both viruses. | SE/leaf disc | Nicotiana tabacum | [29] |
| TuMV + TYMV | HC-Pro (TuMV) + P69 (TYMV) | High protection against both viruses | SE/floral dip | Arabidopsis thaliana | [10] | |||
| 3 | WSMoV | RdRP | High protection | SE/leaf disc | Nicotiana benthamiana | [32] | ||
| AthMIR171a | 2 | TBSV | P19 + P33 | Effective antiviral silencing in agroinfiltrated leaves. | TE/leaf agroinfiltration | Nicotiana benthamiana | [42] | |
| AthMIR319a | 2 | PVYO + PVYN | NIb (PVYO) + NIb (PVYN) | 52% and 30% of the lines were resistant to PVYO and PVYN, respectively. | SE/leaf disc | Nicotiana tabacum | [37] | |
| HvuMIR171a | 3 | WDV | Rep + RepA | One line was fully resistant. | SE/spike immature embryos | Hordeum vulgare, | [40] | |
| Efficient silencing of the overexpressed Rep mRNA at 15ºC and 23ºC. | TE/leaf agroinfiltration | Nicotiana benthamiana | ||||||
| OsaMIR528 | 2 | CymMV + ORSV | RdRP (CymMV) + RdRP (ORSV) | 73% and 16% of the lines were resistant to CymMV and ORSV, respectively. | SE/leaf disc | Nicotiana benthamiana | [43] | |
| OsaMIR528 | 2 | RBSDV + RSV | CP (RBSDV) + CP (RSV) | 54% and 27% of the lines were resistant to RBSDV and RSV, respectively. | SE/scutellum-derived calli | Oryza sativa | [44] | |
| amiRNA/single polycistronic | OsaMIR395 | 5 | WSMV | 5′ UTR + P1 + HC-Pro + P3 | Three types of lines were observed: completely immune; initially resistant with resistance breaking down over time; and initially susceptible followed by plant recovery. | SE/microparticle bombardment of embryos | Triticum aestivum | [45] |
| amiRNA/multiple monocistronic in tandem | AthMIR157 + AthMIR159 + AthMIR171 | 3 | PPV | CP | No protection. | SE/hypocotyl slices | Prunus domestica | [46] |
| amiRNA/multiple monocistronic in trans | AthMIR159a | 2 | TuMV | CP + HC-Pro | High protection. | SE/floral dip | Arabidopsis thaliana | [31] |
| AthMIR390a | 7 | TBSV | 5′ terminus TBSV(+) RNA | 80% of plants were symptom-free. | TE/leaf agroinfiltration | Nicotiana benthamiana | [39] | |
| amiRNA + siRNA/multiple monocistronic in tandem | AthMIR319a + shRNA | 2 | PVYO + PVYN | NIb (PVYO) + NIb (PVYN) | 69% and 47% of the lines were resistant to PVYO and PVYN, respectively. | SE/leaf disc | Nicotiana tabacum | [37] |
| syn-tasiRNA/single polycistronic | AthTAS1c | 1 | TSWV | RdRP | Delay of viroid accumulation. | TE/leaf agroinfiltration | Nicotiana benthamiana | [7] |
| 4 | TSWV | NSm + RdRP | 100% of the plants were resistant. | TE/leaf agroinfiltration | Nicotiana benthamiana, | [18] | ||
| 83% of the lines were resistant. | SE/cotyledonary explants | Solanum lycopersicum | [14] | |||||
| 5 | PSTVd | TL + C + V + TR [PSTVd(+)] + TL + TR [PSTVd(−)] | Delay of viroid accumulation. | TE/leaf agroinfiltration | Nicotiana benthamiana | [17] | ||
| AthTAS3a | 6 | CMV + TuMV | RdRP + 2b (CMV) + P1 + P3 + CP (TuMV) | All lines were resistant to both viruses. | SE/floral dip | Arabidopsis thaliana | [16] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cisneros, A.E.; Carbonell, A. Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants. Plants 2020, 9, 669. https://doi.org/10.3390/plants9060669
Cisneros AE, Carbonell A. Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants. Plants. 2020; 9(6):669. https://doi.org/10.3390/plants9060669
Chicago/Turabian StyleCisneros, Adriana E., and Alberto Carbonell. 2020. "Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants" Plants 9, no. 6: 669. https://doi.org/10.3390/plants9060669
APA StyleCisneros, A. E., & Carbonell, A. (2020). Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants. Plants, 9(6), 669. https://doi.org/10.3390/plants9060669






