Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa Subsp. Pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy
Abstract
:1. Introduction
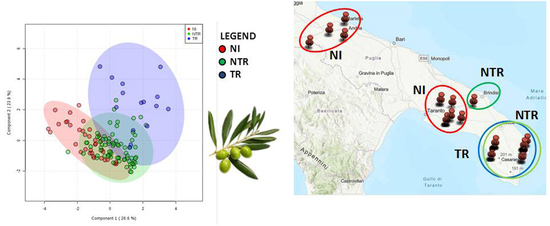
2. Results
2.1. Soil Analyses
2.2. Leaf Analyses
3. Discussion
4. Materials and Methods
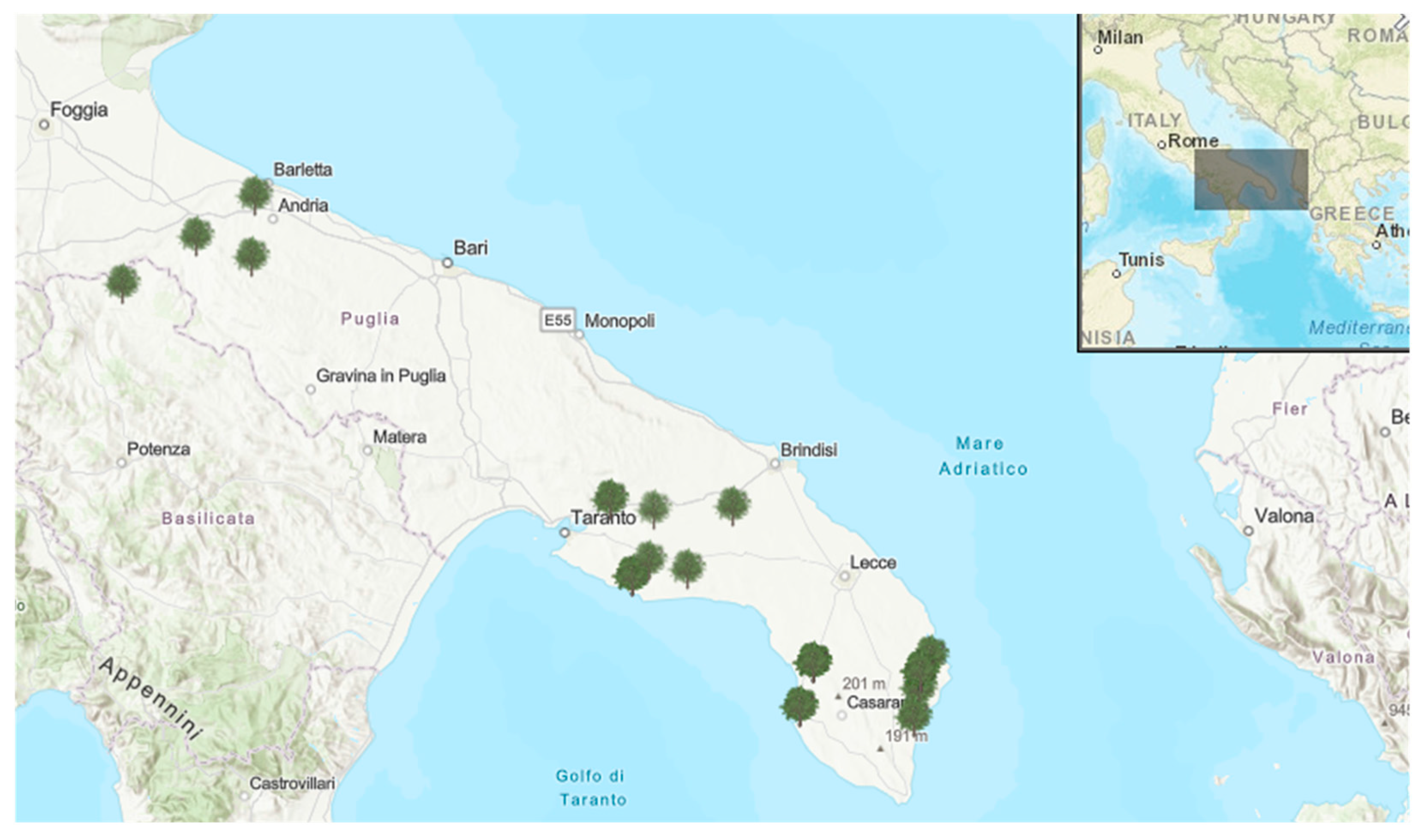
4.1. Soil and Leaf Sampling
4.2. Soil and Leaf Analyses
4.3. Statistical and Principal Component Analyses
4.4. Occurrence of Xylella fastidiosa in Olive Groves
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Leone, A.; Lecci, R.; Durante, M.; Meli, F.; Piraino, S. The bright side of gelatinous blooms: Nutraceutical value and antioxidant properties of three Mediterranean jellyfish (Scyphozoa). Mar. Drugs 2015, 13, 4654–4681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saponari, M.; Boscia, D.; Loconsole, G.; Palmisano, F.; Savino, V.; Potere, O.; Martelli, G. New hosts of Xylella fastidiosa strain CoDiRO in Apulia. J. Plant Pathol. 2014, 96. [Google Scholar] [CrossRef]
- Beck, P. Monitoring the Impact of Xylella on Apulia’s Olive Orchards Using Sentinel-2 Satellite Data and Aerial Photograph. In Proceedings of the 2nd European Conference on Xylella fastidiosa, Ajaccio, Corsica, 29–30 October 2019. [Google Scholar]
- Giampetruzzi, A.; Saponari, M.; Loconsole, G.; Boscia, D.; Savino, V.N.; Almeida, R.P.; Zicca, S.; Landa, B.B.; Chacón-Diaz, C.; Saldarelli, P. Genome-wide analysis provides evidence on the genetic relatedness of the emergent Xylella fastidiosa genotype in Italy to isolates from Central America. Phytopathology 2017, 107, 816–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcelletti, S.; Scortichini, M. Xylella fastidiosa CoDiRO strain associated with the olive quick decline syndrome in southern Italy belongs to a clonal complex of the subspecies pauca that evolved in Central America. Microbiology 2016, 162, 2087–2098. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Saponari, M.; Almeida, R.P.; Essakhi, S.; Boscia, D.; Loconsole, G.; Saldarelli, P. Complete genome sequence of the olive-infecting strain Xylella fastidiosa subsp. pauca De Donno. Genome Announc. 2017, 5, e00569-17. [Google Scholar] [CrossRef] [Green Version]
- Ramazzotti, M.; Cimaglia, F.; Gallo, A.; Ranaldi, F.; Surico, G.; Giovanni, M.; Bleve, G.; Marchi, G. Insights on a founder effect: The case of Xylella fastidiosa in the Salento area of Apulia, Italy. Phytopathol. Mediterr. 2018, 57, 8–25. [Google Scholar]
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Commission Implementing Decision (EU) 2016/764 of 12 May 2016 Amending Implementing Decision (EU) 2015/789 as Regards Measures to Prevent the Introduction into and the Spread within the Union of Xylella Fastidiosa (Wells et al.) (notified under document C(2016) 2731). 2016. Available online: https://op.europa.eu/it/publication-detail/-/publication/7be6a640-199a-11e6-ba9a-01aa75ed71a1/language-en (accessed on 13 June 2020).
- Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.A.; Miret, J.A.J.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P. Update of the Scientific Opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory. EFSA J. 2019, 17, 5665. [Google Scholar]
- Scortichini, M.; Cesari, G. An Evaluation of Monitoring Surveys of the Quarantine Bacterium Xylella Fastidiosa Performed in Containment and Buffer Areas of Apulia, Southern Italy. Appl. Biosaf. 2019, 24, 96–99. [Google Scholar] [CrossRef]
- Cornara, D.; Cavalieri, V.; Dongiovanni, C.; Altamura, G.; Palmisano, F.; Bosco, D.; Porcelli, F.; Almeida, R.; Saponari, M. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. J. Appl. Entomol. 2017, 141, 80–87. [Google Scholar] [CrossRef]
- Martelli, G.P. The current status of the quick decline syndrome of olive in southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Fierro, A.; Liccardo, A.; Porcelli, F. A lattice model to manage the vector and the infection of the Xylella fastidiosa on olive trees. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- White, S.M.; Bullock, J.M.; Hooftman, D.A.; Chapman, D.S. Modelling the spread and control of Xylella fastidiosa in the early stages of invasion in Apulia, Italy. Biol. Invasions 2017, 19, 1825–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strona, G.; Carstens, C.J.; Beck, P.S. Network analysis reveals why Xylella fastidiosa will persist in Europe. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Lahner, B.; Gong, J.; Mahmoudian, M.; Smith, E.L.; Abid, K.B.; Rogers, E.E.; Guerinot, M.L.; Harper, J.F.; Ward, J.M.; McIntyre, L. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 1215–1221. [Google Scholar] [CrossRef]
- Cobine, P.A.; Cruz, L.F.; Navarrete, F.; Duncan, D.; Tygart, M.; De La Fuente, L. Xylella fastidiosa differentially accumulates mineral elements in biofilm and planktonic cells. PLoS ONE 2013, 8, e54936. [Google Scholar] [CrossRef]
- Oliver, J.; Sefick, S.; Parker, J.; Arnold, T.; Cobine, P.; De La Fuente, L. Ionome changes in Xylella fastidiosa–infected Nicotiana tabacum correlate with virulence and discriminate between subspecies of bacterial isolates. Mol. Plant Microbe Interact. 2014, 27, 1048–1058. [Google Scholar] [CrossRef] [Green Version]
- Navarrete, F.; De La Fuente, L. Zinc detoxification is required for full virulence and modification of the host leaf ionome by Xylella fastidiosa. Mol. Plant Microbe Interact. 2015, 28, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Cruz, L.F.; Cobine, P.A.; De La Fuente, L. Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl. Environ. Microbiol. 2012, 78, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- D’Attoma, G.; Morelli, M.; Saldarelli, P.; Saponari, M.; Giampetruzzi, A.; Boscia, D.; Savino, V.N.; De La Fuente, L.; Cobine, P.A. Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of salento, Italy. Pathogens 2019, 8, 272. [Google Scholar] [CrossRef] [Green Version]
- Scortichini, M.; Migoni, D.; Angile, F.; Laura, D.; Girelli, C.R.; Zampella, L.; Mastrobuoni, F.; Fanizzi, F.P. Xylella fastidiosa subsp. pauca on olive in Salento (Southern Italy): Infected trees have low in planta micronutrient content. Phytopathol. Mediterr. 2019, 58, 39–48. [Google Scholar]
- Leite, B.; Ishida, M.; Alves, E.; Carrer, H.; Pascholati, S.; Kitajima, E. Genomics and X-ray microanalysis indicate that Ca2+ and thiols mediate the aggregation and adhesion of Xylella fastidiosa. Braz. J. Med. Biol. Res. 2002, 35, 645–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scortichini, M.; Jianchi, C.; De Caroli, M.; Dalessandro, G.; Pucci, N.; Modesti, V.; L’Aurora, A.; Petriccione, M.; Zampella, L.; Mastrobuoni, F. A zinc, copper and citric acid biocomplex shows promise for control of Xylella fastidiosa subsp. pauca in olive trees in Apulia region (southern Italy). Phytopathol. Mediterr. 2018, 57, 48–72. [Google Scholar]
- Girelli, C.R.; Del Coco, L.; Scortichini, M.; Petriccione, M.; Zampella, L.; Mastrobuoni, F.; Cesari, G.; Bertaccini, A.; D’amico, G.; Contaldo, N. Xylella fastidiosa and olive quick decline syndrome (CoDiRO) in Salento (southern Italy): A chemometric 1H NMR-based preliminary study on Ogliarola salentina and Cellina di Nardò cultivars. Chem. Biol. Technol. Agric. 2017, 4, 25. [Google Scholar] [CrossRef]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR metabolite fingerprinting analysis reveals a disease biomarker and a field treatment response in Xylella fastidiosa subsp. pauca-Infected Olive Trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgson, J. Chemistry of the Micronutrient Elements in Soils. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1963; Volume 15, pp. 119–159. [Google Scholar]
- Sumner, M.E. Sodic soils-New perspectives. Soil Res. 1993, 31, 683–750. [Google Scholar] [CrossRef]
- Alloway, B. Heavy Metals in Soils, 2nd ed.; Chapman and Hall: London, UK, 1995. [Google Scholar]
- McLennan, S.M.; Taylor, S.R. Earth’s Continental Crust. In Encyclopedia of Biochemistry; Marshall, C.P., Fairbridge, R.W., Eds.; Springer: Dordrecht, The Netherlands, 1999. [Google Scholar]
- Kaiser, B.N.; Gridley, K.L.; Ngaire Brady, J.; Phillips, T.; Tyerman, S.D. The role of molybdenum in agricultural plant production. Ann. Bot. 2005, 96, 745–754. [Google Scholar] [CrossRef]
- Kabata, P.; Pendias, H. Trace Elements in Soil and Plants, 4th ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 24–29. [Google Scholar]
- Reimann, C.; Demetriades, A.; Birke, M.; Filzmoser, P.; O’Connor, P.; Halamić, J.; Ladenberger, A.; De Vivo, B.; De Vos, W.; DD, M. Distribution of Elements/Parameters in Agricultural and Grazing Land Soil of Europe. In Chemistry of Europe’s Agricultural Soils—Part A: Methodology and Interoretation of the GEMAS Data Set; Geologische Bundesanstalt für Geowissenschaften und Rohstoffe (BGR): Hannover, Germany, 2014. [Google Scholar]
- Noulas, C.; Tziouvalekas, M.; Karyotis, T. Zinc in soils, water and food crops. J. Trace Elem. Med. Biol. 2018, 49, 252–260. [Google Scholar] [CrossRef]
- Reimann, C.; Birke, M.; Demetriades, A.; Filzmoser, P.; O’Connor, P. Chemistry of Europe’s Agricultural Soils, Part A; Schweizerbarth: Hannover, Germany, 2014. [Google Scholar]
- McDonald, B.A.; Stukenbrock, E.H. Rapid emergence of pathogens in agro-ecosystems: Global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20160026. [Google Scholar] [CrossRef] [Green Version]
- Datnoff, L.E.; Elmer, W.H.; Huber, D.M. Mineral Nutrition and Plant Disease; American Phytopathological Society (APS Press): Saint Paul, MN, USA, 2007. [Google Scholar]
- Eide, D.J. The oxidative stress of zinc deficiency. Metallomics 2011, 3, 1124–1129. [Google Scholar] [CrossRef]
- Caspi, V.; Droppa, M.; Horvath, G.; Malkin, S.; Marder, J.B.; Raskin, V.I. The effect of copper on chlorophyll organization during greening of barley leaves. Photosynth. Res. 1999, 62, 165–174. [Google Scholar] [CrossRef]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in plants: From acquisition to subcellular allocation. Front. Plant Sci. 2020, 11, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabot, C.; Martos, S.; Llugany, M.; Gallego, B.; Toirà, R.; Poschenrieder, C. A role for zinc in plant defense against pathogens and herbivores. Front. Plant Sci. 2019, 10, 1171. [Google Scholar] [CrossRef]
- Kim, S.H.; Hong, J.K.; Lee, S.C.; Sohn, K.H.; Jung, H.W.; Hwang, B.K. CAZFP1, CYS2/HiS(2)-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol. Biol. 2004, 55, 883–904. [Google Scholar] [CrossRef]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Sharma, T.R. Comparative analysis of zinc finger proteins involved in plant disease resistance. PLoS ONE 2012, 7, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poschenrieder, C.; Tolrà, R.; Barceló, J. Can metals defend plants against biotic stress? Trends Plant Sci. 2006, 11, 288–295. [Google Scholar] [CrossRef]
- Fones, H.N.; Preston, G.M. Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. Fems Microbiol. Lett. 2012, 327, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef]
- Martínez-Minaya, J.; Conesa, D.; López-Quílez, A.; Saponari, M.; Vicent, A. Insights into the Spatio-Temporal Spread of Xylella fastidiosa in South-Eastern Italy. In Proceedings of the European Conference on Xylella fastidiosa: Finding Answers to a Global Problem, Palma de Mallorca, Spain, 13–15 November 2017. [Google Scholar]
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). J. Plant Pathol. 2013, 95. [Google Scholar] [CrossRef]
- Andersen, P.; French, W. Biophysical characteristics of peach trees infected with phony peach disease. Physiol. Mol. Plant Pathol. 1987, 31, 25–40. [Google Scholar] [CrossRef]
- Goodwin, P.; DeVay, J.; Meredith, C. Physiological responses of Vitis vinifera cv.“Chardonnay” to infection by the Pierce’s disease bacterium. Physiol. Mol. Plant Pathol. 1988, 32, 17–32. [Google Scholar] [CrossRef]
- De La Fuente, L.; Parker, J.K.; Oliver, J.E.; Granger, S.; Brannen, P.M.; van Santen, E.; Cobine, P.A. The bacterial pathogen Xylella fastidiosa affects the leaf ionome of plant hosts during infection. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson, B. Uptake and translocation of calcium in cucumber. Physiol. Plant. 1982, 54, 107–111. [Google Scholar] [CrossRef]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 2010, 22, 3164–3176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordax, M.; Marco-Noales, E.; López, M.M.; Biosca, E.G. Exopolysaccharides favor the survival of Erwinia amylovora under copper stress through different strategies. Res. Microbiol. 2010, 161, 549–555. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: http://www.R-project.org/ (accessed on 28 February 2020).
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Siepak, M.; Sojka, M. Application of multivariate statistical approach to identify trace elements sources in surface waters: A case study of Kowalskie and Stare Miasto reservoirs, Poland. Environ. Monit. Assess. 2017, 189, 364. [Google Scholar] [CrossRef] [Green Version]
- Harper, S.; Ward, L.; Clover, G. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef] [PubMed]






| BAT-PZ | BR | LE | TA | |
|---|---|---|---|---|
| B | 19.99 ± 0.80 | 16.90 ± 0.71 | 17.36 ± 0.87 | 20.01 ± 1.39 |
| Ca | 44.84 × 103 ± 6.93 × 103 | 31.46 × 103 ± 18.77 × 103 | 68.21 × 103 ± 6.62 × 103 | 80.46 × 103 ± 12.19 × 103 |
| Cu | 84.24 ± 9.30 b | 17.17 ± 4.93 a | 30.78 ± 2.38 a | 20.27 ± 1.84 a |
| Fe | 20.76 × 103 ± 0.98 × 103 | 18.88 × 103 ± 0.70 × 103 | 20.22 × 103 ± 1.03 × 103 | 19.29 × 103 ± 14.16 × 103 |
| Mg | 3.73 × 103 ± 0.14 × 103 b | 2.11 × 103 ± 0.36 × 103 a | 3.21 × 103 ± 0.11 × 103 b | 3.45 × 103 ± 0.14 × 103 b |
| Mn | 0.78 × 103 ± 0.05 × 103 ab | 0.60 × 103 ± 0.08 × 103 ab | 0.46 × 103 ± 0.06 × 103 a | 0.74 × 103 ± 0.079 × 103 b |
| Mo | 0.08 ± 0.02 | 0.24 ± 0.07 | 0.27 ± 0.04 | 0.15 ± 0.03 |
| Na | 3.85 × 103 ± 0.02 × 103c | 0.27 × 103 ± 0.03 × 103 a | 0.62 × 103 ± 0.090 × 103 a | 1.75 × 103 ± 0.24 × 103 b |
| Zn | 46.08 ± 1.74 a | 24.72 ± 1.16 ab | 32.69 ± 2.10 b | 34.71 ± 1.92 ab |
| pH | 8.12 ± 0.04 | 7.64 ± 0.26 | 7.84 ± 0.11 | 8.20 ± 0.04 |
| Ca | Cu | Fe | Mg | Mn | Mo | Zn | Na | |
|---|---|---|---|---|---|---|---|---|
| B | −0.45 *** | 0.13 | 0.89 *** | 0.39 *** | 0.56 *** | 0.02 | 0.52 *** | 0.15 |
| Ca | −0.24 ** | −0.59 *** | 0.31 *** | −0.37 *** | −0.06 | −0.22 | −0.03 | |
| Cu | 0.18 * | 0.09 | 0.16 | 0.14 | 0.38 *** | 0.23 ** | ||
| Fe | 0.32 *** | 0.49 *** | 0.15 | 0.48 *** | 0.04 | |||
| Mg | 0.22 * | 0.03 | 0.38 *** | 0.19 | ||||
| Mn | −0.19 * | 0.46 *** | 0.22 ** | |||||
| Mo | 0.03 | −0.31 ** | ||||||
| Zn | 0.15 |
| Municipality | Province | Status-Cultivar (n° Samples) |
|---|---|---|
| Andria | BAT | NI Coratina (3) |
| Barletta | BAT | NI Coratina (3) |
| Canosa | BAT | NI Coratina (3) |
| Gaudiano | PZ | NI Coratina (3) |
| Francavilla Fontana | BR | NTR Ogliarola (3) |
| Mesagne | BR | NTR Ogliarola (3) |
| Galatone | LE | TR Ogliarola (1)/Cellina (2)-NTR Ogliarola (18) |
| Cannole | LE | TR Cellina (3)-NTR Ogliarola (3) |
| Giurdignano | LE | TR Ogliarola (3)-NTR Ogliarola (3)/Cellina (3) |
| Otranto | LE | TR Ogliarola (3)-NTR Cellina (3) |
| Carpignano | LE | TR Ogliarola (2)/Cellina (1) |
| Ortelle | LE | NTR Cellina (3)- NTR Ogliarola (3) |
| Minervino di Lecce | LE | NTR Cellina (3)- NTR Ogliarola (3) |
| Cursi | LE | NTR Ogliarola (3) |
| Gallipoli | LE | NTR Ogliarola (24) |
| Sava | TA | NI Ogliarola (2)/Leccino (1) |
| Maruggio | TA | NI Ogliarola(1)/Cellina(1)/Leccino(1) |
| Manduria | TA | NI Ogliarola (1)/Cellina (2) |
| Torricella | TA | NI Ogliarola (2)/Leccino (1) |
| Grottaglie | TA | NI Ogliarola (3)/Cellina (3) |
| Lizzano | TA | NI Ogliarola (6) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Coco, L.; Migoni, D.; Girelli, C.R.; Angilè, F.; Scortichini, M.; Fanizzi, F.P. Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa Subsp. Pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy. Plants 2020, 9, 760. https://doi.org/10.3390/plants9060760
Del Coco L, Migoni D, Girelli CR, Angilè F, Scortichini M, Fanizzi FP. Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa Subsp. Pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy. Plants. 2020; 9(6):760. https://doi.org/10.3390/plants9060760
Chicago/Turabian StyleDel Coco, Laura, Danilo Migoni, Chiara Roberta Girelli, Federica Angilè, Marco Scortichini, and Francesco Paolo Fanizzi. 2020. "Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa Subsp. Pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy" Plants 9, no. 6: 760. https://doi.org/10.3390/plants9060760
APA StyleDel Coco, L., Migoni, D., Girelli, C. R., Angilè, F., Scortichini, M., & Fanizzi, F. P. (2020). Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa Subsp. Pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy. Plants, 9(6), 760. https://doi.org/10.3390/plants9060760