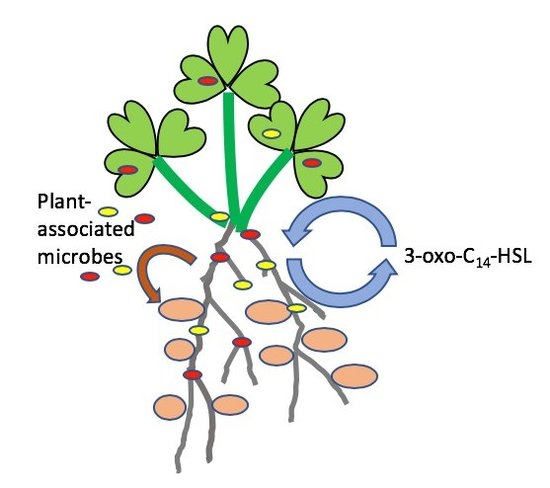
The Presence of Plant-Associated Bacteria Alters Responses to N-acyl Homoserine Lactone Quorum Sensing Signals that Modulate Nodulation in Medicago Truncatula
Abstract
:1. Introduction
2. Results
2.1. The Positive Effect of 3-oxo-C14-HSL on Nodule Numbers Depended on the Presence of Plant-Associated Bacteria
2.2. Interactions of the Effects of the AHL 3-oxo-C14-HSL and the Root Microbiome
3. Discussion
4. Materials and Methods
4.1. Media and Plant Growth Conditions
4.2. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
4.3. Estimation of Bacteria From M. truncatula Roots by Culturing and 16S Amplification
4.4. Bacterial Community Analysis
4.5. Analysis of Microbiome Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Parniske, M. Uptake of bacteria into living plant cells, the unifying and distinct feature of the nitrogen-fixing root nodule symbiosis. Curr. Opin. Plant Biol. 2018, 44, 164–174. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardoim, P.R.; Hardoim, C.C.; van Overbeek, L.S.; van Elsas, J.D. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsek, M.R.; Greenberg, E.P. Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: A signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 2000, 97, 8789–8793. [Google Scholar] [CrossRef] [Green Version]
- Von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Ann. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.; Pierson, E.A.; Pierson, L.S., 3rd; Stacey, G.; Chatterjee, A. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 2002, 5, 285–290. [Google Scholar] [CrossRef]
- Bauer, W.D.; Mathesius, U. Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 2004, 7, 429–433. [Google Scholar] [CrossRef]
- Teplitski, M.; Mathesius, U.; Rumbaugh, K.P. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem. Rev. 2011, 111, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Rothballer, M.; Hense, B.A.; Schroder, P. Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front. Plant Sci. 2014, 5, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anolles, G.; Rolfe, B.G.; Bauer, W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 2003, 100, 1444–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teplitski, M.; Eberhard, A.; Gronquist, M.R.; Gao, M.; Robinson, J.B.; Bauer, W.D. Chemical identification of N-acyl homoserine lactone quorum-sensing signals produced by Sinorhizobium meliloti strains in defined medium. Arch. Microbiol. 2003, 180, 494–497. [Google Scholar] [CrossRef]
- Teplitski, M.; Robinson, J.B.; Bauer, W.D. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe Interact. 2000, 13, 637–648. [Google Scholar] [CrossRef] [Green Version]
- Von Rad, U.; Klein, I.; Dobrev, P.I.; Kottova, J.; Zazimalova, E.; Fekete, A.; Hartmann, A.; Schmitt-Kopplin, P.; Durner, J. Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 2008, 229, 73–85. [Google Scholar] [CrossRef]
- Ortiz-Castro, R.; Martinez-Trujillo, M.; Lopez-Bucio, J. N-acyl-L-homoserine lactones: A class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 2008, 31, 1497–1509. [Google Scholar] [CrossRef]
- Ortiz-Castro, R.; Lopez-Bucio, J. Review: Phytostimulation and root architectural responses to quorum-sensing signals and related molecules from rhizobacteria. Plant Sci. 2019, 284, 135–142. [Google Scholar] [CrossRef]
- Bai, X.; Todd, C.D.; Desikan, R.; Yang, Y.; Hu, X. N-3-oxo-decanoyl-L-homoserine-lactone activates auxin-induced adventitious root formation via hydrogen peroxide- and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol. 2012, 158, 725–736. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Bian, Z.; Jia, Z.; Zhao, Q.; Song, S. The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing signals. Mol. Plant Microbe Interact. 2012, 25, 677–683. [Google Scholar] [CrossRef] [Green Version]
- Zuniga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutierrez, R.A.; Gonzalez, B. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef] [Green Version]
- Schenk, S.T.; Stein, E.; Kogel, K.H.; Schikora, A. Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal. Behav. 2012, 7, 178–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; Van Breusegem, F.; Eberl, L.; et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef]
- Schenk, S.T.; Hernandez-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithofer, A.; Becker, A.; Kogel, K.H.; et al. N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schikora, A.; Schenk, S.T.; Stein, E.; Molitor, A.; Zuccaro, A.; Kogel, K.H. N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 2011, 157, 1407–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrestha, A.; Elhady, A.; Adss, S.; Wehner, G.G.; Boettcher, C.; Heuer, H.; Ordon, F.; Schikora, A. Genetic differences in barley govern the responsiveness to N-acyl homoserine lactone. Phytobiomes J. 2019, 3, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Zarkani, A.A.; Stein, E.; Rohrich, C.R.; Schikora, M.; Evguenieva-Hackenberg, E.; Degenkolb, T.; Vilcinskas, A.; Klug, G.; Kogel, K.H.; Schikora, A. Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 2013, 14, 17122–17146. [Google Scholar] [CrossRef]
- Peters, N.K.; Frost, J.W.; Long, S.R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 1986, 233, 977–980. [Google Scholar] [CrossRef]
- Murray, J.D. Invasion by invitation: Rhizobial infection in legumes. Mol. Plant Microbe Interact. 2011, 24, 631–639. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, G.E.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.P.; Walker, G.C. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 1998, 180, 5183–5191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marketon, M.M.; Gonzalez, J.E. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 2002, 184, 3466–3475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisniewski-Dye, F.; Downie, J.A. Quorum-sensing in Rhizobium. Antonie Van Leeuwenhoek 2002, 81, 397–407. [Google Scholar] [CrossRef]
- González, J.E.; Marketon, M.M. Quorum Sensing in Nitrogen-Fixing Rhizobia. Microbiol. Mol. Biol. Rev. 2003, 67, 574–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Contreras, M.; Bauer, W.D.; Gao, M.S.; Robinson, J.B.; Downie, J.A. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. B 2007, 362, 1149–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Yang, M.; Zheng, H.; Zhang, J.; Zhong, Z.; Zhu, J. Complex quorum-sensing regulatory systems regulate bacterial growth and symbiotic nodulation in Mesorhizobium tianshanense. Arch. Microbiol. 2009, 191, 283–289. [Google Scholar] [CrossRef]
- Mueller, K.; Gonzalez, J.E. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2011, 193, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Coggin, A.; Yagnik, K.; Teplitski, M. Role of specific quorum-sensing signals in the regulation of exopolysaccharide II production within Sinorhizobium meliloti spreading colonies. PLoS ONE 2012, 7, e42611. [Google Scholar] [CrossRef]
- Nievas, F.; Bogino, P.; Sorroche, F.; Giordano, W. Detection, characterization, and biological effect of quorum-sensing signaling molecules in peanut-nodulating bradyrhizobia. Sensors 2012, 12, 2851–2873. [Google Scholar] [CrossRef] [Green Version]
- Veliz-Vallejos, D.F.; van Noorden, G.E.; Yuan, M.; Mathesius, U. A Sinorhizobium meliloti-specific N-acyl homoserine lactone quorum-sensing signal increases nodule numbers in Medicago truncatula independent of autoregulation. Front. Plant Sci. 2014, 5, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.G.; Mukherjee, A.; Stacy, D.M.; Lazar, S.; Ane, J.M.; Blackwell, H.E. Interkingdom responses to bacterial quorum sensing signals regulate frequency and rate of nodulation in legume-rhizobia symbiosis. Chembiochem 2016, 17, 2199–2205. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes 2017, 1, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 1490–1494. [Google Scholar] [CrossRef] [Green Version]
- Marketon, M.M.; Gronquist, M.R.; Eberhard, A.; Gonzalez, J.E. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 2002, 184, 5686–5695. [Google Scholar] [CrossRef] [Green Version]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.; et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Reyes, C.; Schenk, S.T.; Neumann, C.; Kogel, K.-H.; Schikora, A. N-acyl-homoserine lactones-producing bacteria protect plants against plant and human pathogens. Microb. Biotechnol. 2014, 7, 580–588. [Google Scholar]
- Starr, M.P.; Chatterjee, A.K. The genus Erwinia: Enterobacteria pathogenic to plants and animals. Annu. Rev. Microbiol. 1972, 26, 389–426. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda Mdel, C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Brock, A.K.; Berger, B.; Mewis, I.; Ruppel, S. Impact of the PGPB Enterobacter radicincitans DSM 16656 on growth, glucosinolate profile, and immune responses of Arabidopsis thaliana. Microb. Ecol. 2013, 65, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, T.A.; Venter, S.N. Pantoea ananatis: An unconventional plant pathogen. Mol. Plant Pathol. 2009, 10, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Sheibani-Tezerji, R.; Naveed, M.; Jehl, M.A.; Sessitsch, A.; Rattei, T.; Mitter, B. The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front. Microbiol. 2015, 6, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, J.L.; Alvarez, F.; Príncipe, A.; Salas, M.E.; Lozano, M.J.; Draghi, W.O.; Jofré, E.; Lagares, A. Isolation, taxonomic analysis, and phenotypic characterization of bacterial endophytes present in alfalfa (Medicago sativa) seeds. J. Biotechnol. 2018, 267, 55–62. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of plant fraction, soil, and plant species on microbiota: A multikingdom comparison. mBio 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Breidenstein, E.B.; de la Fuente-Nunez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, S.; Wang, J.; Feng, Y. AHL-type quorum sensing and its regulation on symplasmata formation in Pantoea agglomerans YS19. J. Basic Microbiol. 2015, 55, 607–616. [Google Scholar] [CrossRef]
- Chatterjee, A.; Cui, Y.; Hasegawa, H.; Leigh, N.; Dixit, V.; Chatterjee, A.K. Comparative analysis of two classes of quorum-sensing signaling systems that control production of extracellular proteins and secondary metabolites in Erwinia carotovora subspecies. J. Bacteriol. 2005, 187, 8026–8038. [Google Scholar] [CrossRef] [Green Version]
- Lau, Y.Y.; Sulaiman, J.; Chen, J.W.; Yin, W.F.; Chan, K.G. Quorum sensing activity of Enterobacter asburiae isolated from lettuce leaves. Sensors 2013, 13, 14189–14199. [Google Scholar] [CrossRef]
- Dulla, G.; Lindow, S.E. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. USA 2008, 105, 3082–3087. [Google Scholar] [CrossRef] [Green Version]
- Gantner, S.; Schmid, M.; Dürr, C.; Schuhegger, R.; Steidle, A.; Hutzler, P.; Langebartels, C.; Eberl, L.; Hartmann, A.; Dazzo, F.B. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 2006, 56, 188–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, J.E.; Keshavan, N.D. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006, 70, 859–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The yin and yang of bacterial communication. Chembiochem 2009, 10, 205–216. [Google Scholar] [CrossRef]
- Sio, C.F.; Otten, L.G.; Cool, R.H.; Diggle, S.P.; Braun, P.G.; Bos, R.; Daykin, M.; Camara, M.; Williams, P.; Quax, W.J. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 2006, 74, 1673–1682. [Google Scholar] [CrossRef] [Green Version]
- Somers, E.; Vanderleyden, J.; Srinivasan, M. Rhizosphere bacterial signalling: A love parade beneath our feet. Crit. Rev. Microbiol. 2004, 30, 205–240. [Google Scholar] [CrossRef]
- Gao, M.; Chen, H.; Eberhard, A.; Gronquist, M.R.; Robinson, J.B.; Rolfe, B.G.; Bauer, W.D. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2005, 187, 7931–7944. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.; Dreyer, D.; Bonnet, D.; Howell, M.; Nony, E.; VandenBosch, K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 1995, 7, 43–55. [Google Scholar] [PubMed] [Green Version]
- Journet, E.P.; El-Gachtouli, N.; Vernoud, V.; de Billy, F.; Pichon, M.; Dedieu, A.; Arnould, C.; Morandi, D.; Barker, D.G.; Gianinazzi-Pearson, V. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 2001, 14, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, G.E.D.; Downie, J.A. Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 2006, 9, 351–357. [Google Scholar] [CrossRef]
- Götz-Rösch, C.; Sieper, T.; Fekete, A.; Schmitt-Kopplin, P.; Hartmann, A.; Schröder, P. Influence of bacterial N-acyl-homoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front. Plant Sci. 2015, 6, 205. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Jia, Z.; Xu, J.; Zhang, Z.; Bian, Z. N-butyryl-homoserine lactone, a bacterial quorum-sensing signaling molecule, induces intracellular calcium elevation in Arabidopsis root cells. Biochem. Biophys. Res. Commun. 2011, 414, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G.; Senechal, A.C.; Mukherjee, A.; Ané, J.M.; Blackwell, H.E. Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem. Biol. 2014, 9, 1834–1845. [Google Scholar] [CrossRef] [Green Version]
- White, L.J.; Jothibasu, K.; Reese, R.N.; Brozel, V.S.; Subramanian, S. Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol. Plant Microbe Interact. 2015, 28, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar] [CrossRef] [Green Version]
- Fahraeus, G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957, 16, 374–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, E.W.; Brown, A.B.; Nesnas, N.; Palmer, A.G. Abiotic hydrolysis kinetics of N-acyl-L-homoserine lactones: Natural silencing of bacterial quorum sensing signals. Eur. J. Org. Chem. 2019, 2019, 2850–2856. [Google Scholar] [CrossRef]
- Rolfe, B.G.; Gresshoff, P.M.; Shine, J. Rapid screening for symbiotic mutants of Rhizobium and white clover. Plant Sci. Lett. 1980, 19, 277–284. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [Green Version]
- Kakar, K.; Wandrey, M.; Czechowski, T.; Gaertner, T.; Scheible, W.R.; Stitt, M.; Torres-Jerez, I.; Xiao, Y.; Redman, J.C.; Wu, H.C.; et al. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 2008, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelius, M.K.; Triplett, E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowd, S.E.; Sun, Y.; Wolcott, R.D.; Domingo, A.; Carroll, J.A. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: Bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Path. Dis. 2008, 5, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Eren, A.M.; Zozaya, M.; Taylor, C.M.; Dowd, S.E.; Martin, D.H.; Ferris, M.J. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS ONE 2011, 6, e26732. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef] [Green Version]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011, 5, 639–649. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 22 June 2020).
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R package version 2.5-6. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html/ (accessed on 22 June 2020).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]










| Gene Name | Primer Sequence | Reference |
|---|---|---|
| GAPDHF | 5′ TGCCTACCGTCGATGTTTCAGT 3′ | [80] |
| GAPDHR | 5′ TTGCCCTCTGATTCCTCCTTG 3′ | [80] |
| ERN1F | 5′ TGCATGCCTTCTTCGAGGTTCG 3′ | This study |
| ERN1R | 5′ TCCTGGAAGCAAGAGGAGAATCC 3′ | This study |
| RIP1F | 5′ GCTAGATGATACCCCAAATTTCA 3′ | This study |
| RIP1R | 5′ CCACAGAAAATCCTCTGATTGA 3′ | This study |
| MtNIN1F | 5′ GGGAGAAAGTCCGGGGACAA 3′ | [78] |
| MtNIN1R | 5′ GACACACACCGATGCTCTTTGC 3′ | [78] |
| ENOD11F | 5′ TATGGTAACCAGCCTCCACCTAGC 3′ | This study |
| ENOD11R | 5′ GCATTGGTAAACCTTGTTGCTTGC 3′ | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veliz-Vallejos, D.F.; Kawasaki, A.; Mathesius, U. The Presence of Plant-Associated Bacteria Alters Responses to N-acyl Homoserine Lactone Quorum Sensing Signals that Modulate Nodulation in Medicago Truncatula. Plants 2020, 9, 777. https://doi.org/10.3390/plants9060777
Veliz-Vallejos DF, Kawasaki A, Mathesius U. The Presence of Plant-Associated Bacteria Alters Responses to N-acyl Homoserine Lactone Quorum Sensing Signals that Modulate Nodulation in Medicago Truncatula. Plants. 2020; 9(6):777. https://doi.org/10.3390/plants9060777
Chicago/Turabian StyleVeliz-Vallejos, Debora F., Akitomo Kawasaki, and Ulrike Mathesius. 2020. "The Presence of Plant-Associated Bacteria Alters Responses to N-acyl Homoserine Lactone Quorum Sensing Signals that Modulate Nodulation in Medicago Truncatula" Plants 9, no. 6: 777. https://doi.org/10.3390/plants9060777
APA StyleVeliz-Vallejos, D. F., Kawasaki, A., & Mathesius, U. (2020). The Presence of Plant-Associated Bacteria Alters Responses to N-acyl Homoserine Lactone Quorum Sensing Signals that Modulate Nodulation in Medicago Truncatula. Plants, 9(6), 777. https://doi.org/10.3390/plants9060777