Exogenous Carbon Compounds Modulate Tomato Root Development
Abstract
:1. Introduction
2. Results
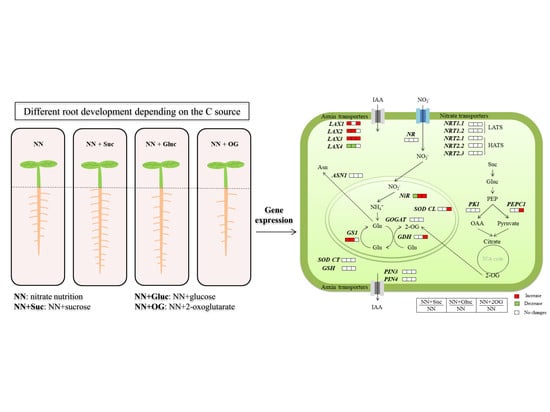
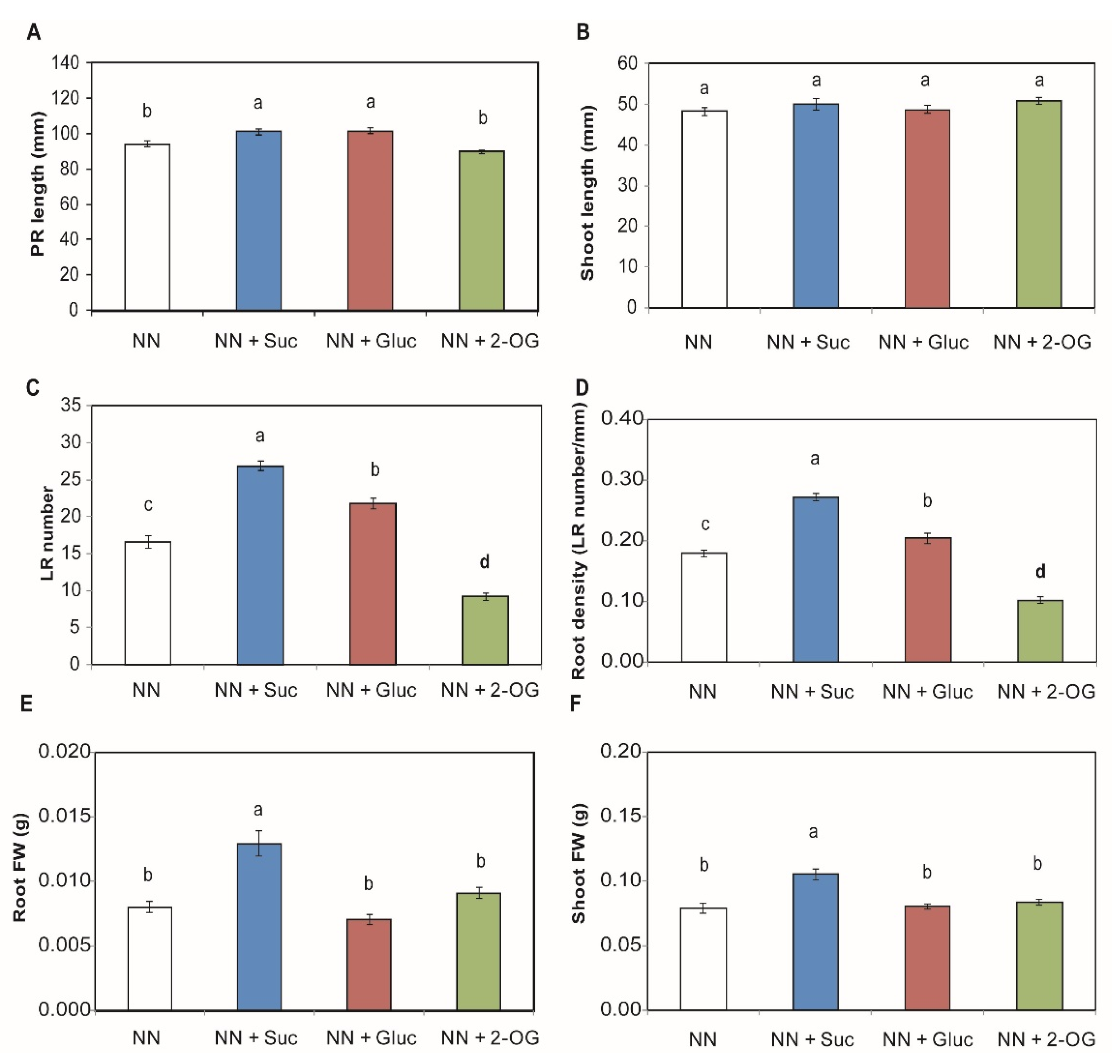
2.1. Carbon Sources Effect on Root and Shoot Development
2.2. Carbon Sources Effect on NO3− Assimilation
2.3. Auxin Transporters Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Root and Shoot Measurements
4.3. qRT-PCR Analyses
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alvarez, J.M.; Vidal, E.; Gutiérrez, R.A. Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 2012, 15, 185–191. [Google Scholar] [CrossRef]
- Crawford, N.; Forde, B. Molecular and Developmental Biology of Inorganic Nitrogen Nutrition. Arab. Book 2002, 1, e0011. [Google Scholar] [CrossRef] [Green Version]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, P. Dancing with Hormones: A Current Perspective of Nitrate Signaling and Regulation in Arabidopsis. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Foehse, D.; Jungk, A. Influence of phosphate and nitrate supply on root hair formation of rape, spinach and tomato plants. Plant Soil 1983, 74, 359–368. [Google Scholar] [CrossRef]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Von Wirén, N. Signaling pathways underlying nitrogen-dependent changes in root system architecture: From model to crop species. J. Exp. Bot. 2020. [Google Scholar] [CrossRef]
- Willaume, M.; Pagès, L. How periodic growth pattern and source/sink relations affect root growth in oak tree seedlings. J. Exp. Bot. 2006, 57, 815–826. [Google Scholar] [CrossRef]
- Mishra, B.S.; Singh, M.; Aggrawal, P.; Laxmi, A. Glucose and Auxin Signaling Interaction in Controlling Arabidopsis thaliana Seedlings Root Growth and Development. PLoS ONE 2009, 4, e4502. [Google Scholar] [CrossRef]
- Mudgil, Y.; Karve, A.; Teixeira, P.J.; Jiang, K.; Tunc-Ozdemir, M.; Jones, A.M. Photosynthate Regulation of the Root System Architecture Mediated by the Heterotrimeric G Protein Complex in Arabidopsis. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Roycewicz, P.; Malamy, J. Dissecting the effects of nitrate, sucrose and osmotic potential on Arabidopsis root and shoot system growth in laboratory assays. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1489–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Zhang, J.; Gao, W.; Chen, Y.; Li, H.; Lawlor, D.W.; Paul, M.J.; Pan, W. Exogenous trehalose improves growth under limiting nitrogen through upregulation of nitrogen metabolism. BMC Plant Biol. 2017, 17, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, A.J.; Cramer, M.D. Root Nitrogen Acquisition and Assimilation. Plant Soil 2005, 274, 1–36. [Google Scholar] [CrossRef]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.T. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Hildebrandt, U.; Schmelzer, E.; Bothe, H. Expression of nitrate transporter genes in tomato colonized by an arbuscular mycorrhizal fungus. Physiol. Plant. 2002, 115, 125–136. [Google Scholar] [CrossRef]
- Lauter, F.R.; Ninnemann, O.; Bucher, M.; Riesmeier, J.W.; Frommer, W.B. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 8139–8144. [Google Scholar] [CrossRef] [Green Version]
- Ono, F.; Frommer, W.B.; Von Wirén, N. Coordinated Diurnal Regulation of Low- and High-Affinity Nitrate Transporters in Tomato. Plant Boil. 2000, 2, 17–23. [Google Scholar] [CrossRef]
- Miflin, B.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [Green Version]
- Herrera-Rodríguez, M.B.; Carrasco-Ballesteros, S.; Maldonado, J.M.; Pineda, M.; Aguilar, M.; Pérez-Vicente, R. Three genes showing distinct regulatory patterns encode the asparagine synthetase of sunflower (Helianthus annuus). New Phytol. 2002, 155, 33–45. [Google Scholar] [CrossRef]
- Forde, B.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Krapp, A. The interaction between elevated carbon dioxide and nitrogen nutrition: The physiological and molecular background. Plant Cell Environ. 1999, 22, 583–621. [Google Scholar] [CrossRef]
- Martin, T.; Oswald, O.; Graham, I.A. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 2002, 128, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E.; Ryan, K.S. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001, 127, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Rong, H.; Pilbeam, D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 2329–2338. [Google Scholar] [CrossRef] [Green Version]
- Blakeslee, J.J.; Peer, W.A.; Murphy, A.S. Auxin transport. Curr. Opin. Plant Biol. 2005, 8, 494–500. [Google Scholar] [CrossRef]
- Geisler, M.; Murphy, A.S. The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 2005, 580, 1094–1102. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.-H.; Yu, J.-Q.; Hu, D.-G. Nitrate: A Crucial Signal during Lateral Roots Development. Front. Plant Sci. 2017, 8, 2029. [Google Scholar] [CrossRef]
- Swarup, K.; Benkova, E.; Swarup, R.; Casimiro, I.; Peret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Pattison, R.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef] [PubMed]
- Peret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.K.; et al. AUX/LAX Genes Encode a Family of Auxin Influx Transporters That Perform Distinct Functions during Arabidopsis Development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, E.; Gutiérrez, R.A. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 2008, 11, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Ivanchenko, M.G.; Os, D.D.; Monshausen, G.B.; Dubrovsky, J.G.; Bednarova, A.; Krishnan, N. Auxin increases the hydrogen peroxide (H2O2) concentration in tomato (Solanum lycopersicum) root tips while inhibiting root growth. Ann. Bot. 2013, 112, 1107–1116. [Google Scholar] [CrossRef] [Green Version]
- Mangano, S.; Denita-Juarez, S.P.; Choi, H.-S.; Marzol, E.; Hwang, Y.; Ranocha, P.; Velasquez, S.M.; Borassi, C.; Barberini, M.L.; Aptekmann, A.A.; et al. Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. USA 2017, 114, 5289–5294. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Tsukagoshi, H. Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol. 2016, 29, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Manzano, C.; Pallero-Baena, M.; Casimiro, I.; De Rybel, B.; Orman-Ligeza, B.; Van Isterdael, G.; Beeckman, T.; Draye, X.; Casero, P.; Del Pozo, J.C. The Emerging Role of Reactive Oxygen Species Signaling during Lateral Root Development. Plant Physiol. 2014, 165, 1105–1119. [Google Scholar] [CrossRef] [Green Version]
- Keunen, E.; Peshev, D.; Vangronsveld, J.; Ende, W.V.D.; Cuypers, A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 2013, 36, 1242–1255. [Google Scholar] [CrossRef]
- Garcia, B.M.; Njo, M.; Beeckman, T.; Goormachtig, S.; Foyer, C.H. A new role for glutathione in the regulation of root architecture linked to strigolactones. Plant Cell Environ. 2013, 37, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Schnaubelt, D.; Queval, G.; Dong, Y.; Diaz-Vivancos, P.; Makgopa, M.; Howell, G.; De Simone, A.; Bai, J.; Hannah, M.A.; Foyer, C.H. Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol inArabidopsis thaliana. Plant Cell Environ. 2014, 38, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Lehmann, M.; Schwarzländer, M.; Baxter, C.J.; Sienkiewicz-Porzucek, A.; Williams, T.C.; Schauer, N.; Fernie, A.R.; Fricker, M.; Ratcliffe, G.; et al. Decrease in Manganese Superoxide Dismutase Leads to Reduced Root Growth and Affects Tricarboxylic Acid Cycle Flux and Mitochondrial Redox Homeostasis. Plant Physiol. 2008, 147, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Malamy, J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef]
- Freixes, S.; Thibaud, M.-C.; Tardieu, F.; Muller, B. Root elongation and branching is related to local hexose concentration in Arabidopsis thaliana seedlings. Plant Cell Environ. 2002, 25, 1357–1366. [Google Scholar] [CrossRef]
- Lee-Ho, E.; Walton, L.J.; Reid, D.M.; Yeung, E.C.; Kurepin, L.V. Effects of elevated carbon dioxide and sucrose concentrations on Arabidopsis thaliana root architecture and anatomy. Can. J. Bot. 2007, 85, 324–330. [Google Scholar] [CrossRef]
- MacGregor, D.R.; Deak, K.I.; Ingram, P.A.; Malamy, J. Root System Architecture in Arabidopsis Grown in Culture Is Regulated by Sucrose Uptake in the Aerial Tissues. Plant Cell 2008, 20, 2643–2660. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.; Gamage, D.; Hirotsu, N.; Martin, A.; Seneweera, S. Effects of Elevated Carbon Dioxide on Photosynthesis and Carbon Partitioning: A Perspective on Root Sugar Sensing and Hormonal Crosstalk. Front. Physiol. 2017, 8, 578. [Google Scholar] [CrossRef] [Green Version]
- Walch-Liu, P.; Ivanov, I.I.; Filleur, S.; Gan, Y.; Remans, T.; Forde, B. Nitrogen Regulation of Root Branching. Ann. Bot. 2005, 97, 875–881. [Google Scholar] [CrossRef]
- González-Hernández, A.I.; Scalschi, L.; García-Agustín, P.; Camañes, G. Tomato root development and N assimilation depend on C and ABA content under different N sources. Plant Physiol. Biochem. 2020, 148, 368–378. [Google Scholar] [CrossRef]
- Lejay, L.; Gansel, X.; Cerezo, M.; Tillard, P.; Müller, C.; Krapp, A.; Von Wirén, N.; Daniel-Vedele, F.; Gojon, A. Regulation of Root Ion Transporters by Photosynthesis: Functional Importance and Relation with Hexokinase. Plant Cell 2003, 15, 2218–2232. [Google Scholar] [CrossRef] [Green Version]
- Lejay, L.; Wirth, J.; Pervent, M.; Cross, J.; Tillard, P.; Gojon, A. Oxidative Pentose Phosphate Pathway-Dependent Sugar Sensing as a Mechanism for Regulation of Root Ion Transporters by Photosynthesis. Plant Physiol. 2008, 146, 2036–2053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, F.; Thodey, K.; Lejay, L.V.; Bevan, M.W. Glucose elevates NITRATE TRANSPORTER2.1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2.1 expression. Plant Physiol. 2013, 164, 308–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [PubMed] [Green Version]
- Lillo, C.; Appenroth, K.J. Light Regulation of Nitrate Reductase in Higher Plants: Which Photoreceptors are Involved? Plant Biol. 2001, 3, 455–465. [Google Scholar] [CrossRef]
- Ali, A.; Sivakami, S.; Raghuram, N. Effect of nitrate, nitrite, ammonium, glutamate, glutamine and 2-oxoglutarate on the RNA levels and enzyme activities of nitrate reductase and nitrite reductase in rice. Physiol. Mol. Biol. Plants 2007, 13, 17–25. [Google Scholar]
- Guan, M.; De Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic Glutamine Synthetase Gln1;2 Is the Main Isozyme Contributing to GS1 Activity and Can Be Up-Regulated to Relieve Ammonium Toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef] [Green Version]
- Hachiya, T.; Sakakibara, H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 2016, 68, 2501–2512. [Google Scholar] [CrossRef]
- Price, J.; Laxmi, A.; Martin, S.K.S.; Jang, J.-C. Global Transcription Profiling Reveals Multiple Sugar Signal Transduction Mechanisms in Arabidopsis. Plant Cell 2004, 16, 2128–2150. [Google Scholar] [CrossRef] [Green Version]
- Setién, I.; Vega-Mas, I.; Celestino, N.; Calleja-Cervantes, M.E.; González-Murua, C.; Estavillo, J.-M.; González-Moro, M.B. Root phosphoenolpyruvate carboxylase and NAD-malic enzymes activity increase the ammonium-assimilating capacity in tomato. J. Plant Physiol. 2014, 171, 49–63. [Google Scholar] [CrossRef]
- Laskowski, M.; Grieneisen, V.A.; Hofhuis, H.; Hove, C.A.T.; Hogeweg, P.; Marée, A.F.; Scheres, B. Root System Architecture from Coupling Cell Shape to Auxin Transport. PLoS Biol. 2008, 6, e307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, R.; Bhosale, R. Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef] [Green Version]
- Sairanen, I.; Novak, O.; Pěnčík, A.; Ikeda, Y.; Jones, B.; Sandberg, G.; Ljung, K. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 2012, 24, 4907–4916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revalska, M.; Vassileva, V.; Zechirov, G.; Iantcheva, A. Is the auxin influx carrier LAX3 essential for plant growth and development in the model plants Medicago truncatula, Lotus japonicus and Arabidopsis thaliana? Biotech. Biotechnol. Equip. 2015, 29, 786–797. [Google Scholar] [CrossRef]
- Bao, Y.; Huang, X.; Rehman, M.; Wang, Y.; Wang, B.; Peng, D. Identification and Expression Analysis of the PIN and AUX/LAX Gene Families in Ramie (Boehmeria nivea L. Gaud). Agron 2019, 9, 435. [Google Scholar] [CrossRef] [Green Version]
- Koch, K.E. Carbohydrate-Modulated Gene Expression in Plants. Annu. Rev. Plant Biol. 1996, 47, 509–540. [Google Scholar] [CrossRef] [Green Version]
- González-Hernández, A.I.; Fernández-Crespo, E.; Scalschi, L.; Hajirezaei, M.-R.; Von Wirén, N.; García-Agustín, P.; Camañes, G. Ammonium mediated changes in carbon and nitrogen metabolisms induce resistance against Pseudomonas syringae in tomato plants. J. Plant Physiol. 2019, 239, 28–37. [Google Scholar] [CrossRef]
- Seifi, H.S.; Curvers, K.; De Vleesschauwer, D.; Delaere, I.; Aziz, A.; Höfte, M. Concurrent overactivation of the cytosolic glutamine synthetase and the GABA shunt in the ABA-deficientsitiensmutant of tomato leads to resistance againstBotrytis cinerea. New Phytol. 2013, 199, 490–504. [Google Scholar] [CrossRef]
- Yao, J.; Shi, W.; Xu, W.F. Effects of salt stress on expression of nitrate transporter and assimilation-related genes in tomato roots. Russ. J. Plant Physiol. 2008, 55, 232–240. [Google Scholar] [CrossRef]
- Yin, Y.-G.; Tominaga, T.; Iijima, Y.; Aoki, K.; Shibata, D.; Ashihara, H.; Nishimura, S.; Ezura, H.; Matsukura, C. Metabolic Alterations in Organic Acids and γ-Aminobutyric Acid in Developing Tomato (Solanum lycopersicum L.) Fruits. Plant Cell Physiol. 2010, 51, 1300–1314. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Hernández, A.I.; Scalschi, L.; García-Agustín, P.; Camañes, G. Exogenous Carbon Compounds Modulate Tomato Root Development. Plants 2020, 9, 837. https://doi.org/10.3390/plants9070837
González-Hernández AI, Scalschi L, García-Agustín P, Camañes G. Exogenous Carbon Compounds Modulate Tomato Root Development. Plants. 2020; 9(7):837. https://doi.org/10.3390/plants9070837
Chicago/Turabian StyleGonzález-Hernández, Ana Isabel, Loredana Scalschi, Pilar García-Agustín, and Gemma Camañes. 2020. "Exogenous Carbon Compounds Modulate Tomato Root Development" Plants 9, no. 7: 837. https://doi.org/10.3390/plants9070837
APA StyleGonzález-Hernández, A. I., Scalschi, L., García-Agustín, P., & Camañes, G. (2020). Exogenous Carbon Compounds Modulate Tomato Root Development. Plants, 9(7), 837. https://doi.org/10.3390/plants9070837